All published articles of this journal are available on ScienceDirect.
Ancestry of Amerindians and its Impact in Anthropology, Transplantation, HLA Pharmacogenomics and Epidemiology by HLA Study in Wiwa Colombian Population
Abstract
Background:
HLA autosomic genes are unique because they conform the most polymorphic human system. Importance of this system is recognized in Medicine for Transplantation, Epidemiology (HLA and disease linkage), Pharmacogenomics (HLA linked to drug side effects) and for defining the origin of populations in both male and female lineages.
Objectives:
Studying HLA profile of a isolated Amerindian group from North Colombia (Wiwa) in order to draw conclusions about its Preventive Medicine, the genetic relationship with Worldwide populations and America peopling, since this last issue is hotly debated.
Methodology:
A total of 14,660 HLA chromosomes were included. Peripheral blood was obtained from volunteer blood donors belonging to Wiwa (also named Arsario) ethnic group. HLA-A, -B, -C, -DRB1 and -DQB1 genes were analyzed by standard methods. Wiwa Amerindians relationships with others were calculated by using Arlequin, Dispan and Vista software computer packages.
Results:
Extended HLA, -A, -B, -C, -DRB1 and -DQB1 haplotypes have been studied for the first time in this population. Classical Amerindian haplotypes have been found and also new Wiwa (Arsario) Amerindian haplotypes. New haplotypes are A*68:01 - B*15:01 - C*03:03 - DRB1*14:02 - DQB1*03:02, A*11:01 - B*07:02 - C*07:02 - DRB1*15:03 - DQB1*06:02 and A*68:01 - B*15:01 - C*03:04 - DRB1*14:02 - DQB1*03:01.
Conclusion:
They have been reached after exhaustive comparisons of Wiwa with other Amerindians and Worldwide populations by using genetic distances, Neighbor Joining trees, correspondence analysis and specific group of alleles which are common and frequent in both Amerindians and Pacific Islanders. They are: 1) The Americas First Inhabitants have been probably come through Bering Strait and also through Pacific (from Austronesia and Asia) and Atlantic (from Europe) routes. A bidirectional gene flow is not discarded. 2) Genetic HLA Amerindian profile is separated from that of other Worldwide populations. 3) Amerindians geographical proximity groups’ relatedness is not concordant with HLA genetic relatedness, neither with language. This may be explained by a substantial population decrease that occurred after Europeans invaded America in 1492 and carried new pathogens and epidemics. 4) Our results are also useful for Wiwa and other Amerindians future preventive medicine (HLA linked diseases), HLA pharmacogenomics and transplantation regional programs.
INTRODUCTION
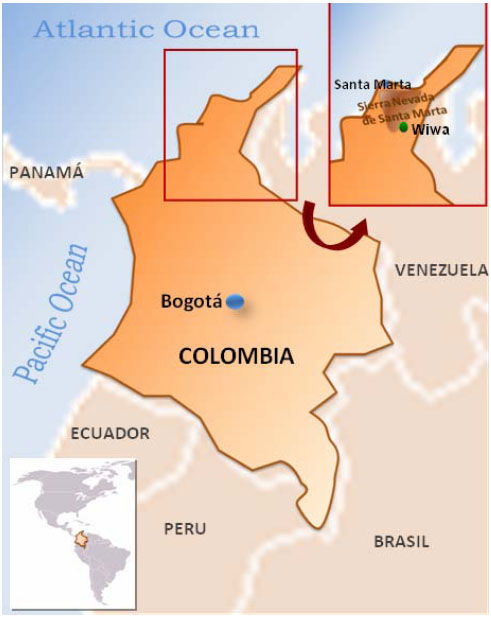
Wiwa ethnic group is located in the northern part of Colombia, in the departments of Cesar, La Guajira and Magdalena. Some of them inhabit at Sierra Nevada de Santa Marta mountains and some of their more important settlements are located between the upper middle basin of the Rancheira river at the Cesar and Badillo rivers (Fig. 1). Their mother language is Damana that belongs to the Chibcha speaking group of languages [1].
First Amerindians are thought to have come from Asia through the Bering land bridge that existed between 30,000-12,000 years before present (BP). These conclusions were reached on a cultural, morphological and genetic background which is common to American and Asian populations. Both Siberia [2] and Mongolia [3, 4] have been put forward as Asia origin. Greenberg postulated a three wave migration for the peopling of the Americas [5]: Amerindians (most North and South American Indians; 12,000 years BP), Na-Dene (Athabaskans, Navajo, Apache; 8,000 years BP) and Eskimo-Aleuts (6,000 years BP). Research conducted with use of Y Chromosome (Y Chr) and other nuclear DNA markers [6-8] supported the three-wave model. However, other mtDNA studies have not [9, 10]; others also postulate only one wave coming from Asia as giving rise to the First Native American ancestors [3, 4].
Also, Amerindians entrance from Asia may have occurred through Coastal Pacific line [11]. A Trans-Pacific route of people from Asia or Polynesia [12] has been suggested because HTLV-1 virus strains shared identical traits both in Japan and South America [13] and certain HLA alleles may have been introduced by the same route [14, 15]. In the same way, “quasi-specific” Amerindian HLA alleles, like A*02:12 or B*39:05 [16], have been found in unrelated individuals of Easter Island, which implies an early contact between Easter Islanders Polynesians and Amerindians [17]. Genetic studies have identified Polynesian mtDNA haplogroups in remains (skulls) of Botocudo Amerindians from Brazil (Amerindian group extinct by the end of 19th century) [18]. Other cultural traits indicate a communication between these groups of populations, i.e.: the presence of South American sweet potato in earlier Pacific sites [19] and the finding of chicken remains of Polynesian type in El Arenal (Chile) dated by radiocarbon back to 1300-1400 AD [20, 21]. In addition, skeletal remains of pre-Columbian individuals with Polynesian ancestry and several Mapuche (Chilean Amerindians) artefacts which are similar to Polynesian ones at Mocha island (Chile) have also been reported [19, 22]. All these facts provide evidences for this Trans-Pacific route that could have occurred in both ways at different times.
Indeed, we have already concluded from our previous studies on HLA genes that: 1 -Aleuts are a genetic and linguistic separate group, possibly related to northern European Lapps; both of them came from southern Siberia Baikal Lake area. 2- First America Natives, which include all analyzed Amerindians, Na-Dene speakers and Eskimo have had genetic flow with Pacific Islanders: the latter share autosomal HLA alleles and haplotypes with them. This fact may have been bidirectional. 3- More important, Easter Islanders show a probable cultural and genetic exchange with Titikaka Lake Aymaras; this civilisation also shares specific traits with European-Iberian megalithic builders. 4- Mesoamericans are grouped together because of they bear more ancient Olmec culture traits and present paper HLA results. 5- Genetics is not able by itself to find out in space and time Americas peopling and First America Inhabitants relatedness with Pacific Islanders [12].
Finally, both genetic [23] and archaeological [24] findings indicate that a two-way Trans-Atlantic traffic may have occurred before Columbus discovered America; archaeologists in New Mexico and other North America places have recently found tools used 20,000 years ago in Spanish Solutrean culture [24, 25].
In the present work, we have studied the Wiwa ethnic group HLA allele frequencies and have compared them with those of Caucasian European, Siberians, Orientals, Na-Dene, Eskimos and Amerindian populations. HLA genes have been analyzed for the following Amerindian ethnic groups: Mayans [26], Mixe, Mixtecans, Zapotecans [27], Lakota Sioux [28], Mazatecans [29], Lamas [30], Quechuas [31], Aymaras [16], Uros [14], Tarahumaras [32], Mapuches [33], Toba Pilaga, Mataco Wichi, and Eastern Toba [15].
Our aims are: 1) To determine the HLA class I (A, B and C) and HLA class II (DRB1 and DQB1) quasi-specific Wiwa Amerindian alleles and specific HLA haplotypes by using indirect DNA sequencing; in other words, the most frequent HLA alleles and haplotypes in this ethnic group which are not found or are in very low frequency in other populations, i.e.: genealogy comparisons, 2) To compare the Wiwa Amerindians HLA allele frequencies with those of other First Americans Native (Na- Dene, Eskimo and Aleuts) and also those of other Asian and Pacific populations (Table 1) in order to find out the HLA relatedness with peoples most likely to be candidates for First Americans ancestors, i.e.: groups of genes frequencies comparisons by using class I and/or II HLA alleles and haplotypes frequencies. 3) To studying specific HLA alleles that are very frequent in Pacific Ocean people and common to both Pacific Ocean people and our Wiwa sample and 4) To establish the Wiwa profile that will be useful for preventive HLA genetic Epidemiology (HLA linked diseases), HLA Pharmacogenomics and future Transplantation waiting lists.
| Population | N | References | Population | N | References |
|---|---|---|---|---|---|
| Wiwa | 55 | Present Study | Evenks | 35 | [36] |
| Mazatecans | 89 | [29] | Aleuts | 85 | [37] |
| Mayans | 132 | [26] | Singapore Chinese | 71 | [38] |
| Aymara | 102 | [16] | Buyi | 70 | [38] |
| Lamas | 83 | [30] | Manchu | 50 | [38] |
| Quechuas | 80 | [31] | Koreans | 100 | [38] |
| Seri | 100 | [27] | Japanese | 493 | [38] |
| Mixe | 55 | [27] | Khalk Mongolians | 202 | [39] |
| Mixtecans | 103 | [27] | Tuvinians | 197 | [40] |
| Zapotecans | 75 | [27] | Khoton Mongolians | 85 | [39] |
| Mexican Mestizo | 99 | Unpublished Results | Germans | 295 | [38] |
| Wayu | 58 | [41] | Danish | 124 | [38] |
| Arhuaco | 123 | [42] | Sardinians | 91 | [38] |
| Kogi | 67 | [42] | Italians | 184 | [38] |
| Mapuches | 104 | [33] | French | 179 | [38] |
| Cayapa | 100 | [43] | Spaniards | 176 | [44] |
| Xavantes | 74 | [15] | Spanish Basques | 80 | [44] |
| Guarani | 32 | [27] | Algerians | 102 | [45] |
| Toba-Pilaga | 19 | [15] | Berbers (Souss) | 98 | [46] |
| Mataco-Wichi | 49 | [15] | Macedonians | 172 | [47] |
| Eastern Toba | 135 | [15] | Cretans | 135 | [48] |
| Jaiduakama | 39 | [49] | Bushmen | 77 | [38] |
| Tarahumara | 44 | [32] | South African Blacks | 86 | [38] |
| Nahuas | 85 | [50] | North American Blacks | 132 | [38] |
| Mayos | 60 | [51] | Chuvashians | 82 | [52] |
| Teenek | 55 | [53] | Russians | 200 | [54] |
| Terena | 60 | [55] | Western Samoa | 102 | [56] |
| Uros | 105 | [14] | Madang | 65 | [57] |
| Lakota Sioux | 302 | [28] | Rabaul | 60 | [57] |
| Eskimos | 35 | [36] | New Caledonia | 65 | [57] |
| Athabascans | 124 | [58] | Fidji | 57 | [57] |
| Nivkhs | 32 | [36] | Papua New Guinea | 57 | [57] |
| Tlinglit | 53 | [38] | Central Desert | 152 | [59] |
| Udegeys | 23 | [36] | Ainu | 50 | [60] |
| Koryaks | 92 | [36] | Yuendumu | 119 | [59] |
| Chukchi | 59 | [36] | Cape York | 80 | [61] |
| Kets | 22 | [36] | Kimberley | 82 | [61] |
MATERIAL AND METHODS
Population Sample
55 healthy unrelated volunteers from the Wiwa ethnic group were HLA class I and class II typed. The Wiwa communities are inhabitants of the Sierra Nevada de Santa Marta in the northern part of Colombia (departments of Magdalena and Cesar). Samples were taken from unrelated individuals from “El Encanto” town at Guanachaca riverside (see map Fig. 1). A writing consent to participate in the present study was signed by each individual. Each individual was born in this area, had a Wiwa physical appearance and their four grandparents had been born in the same area and spoke their own language. We compare our data with those of Caucasian Europeans, Siberians, Orientals, Na-Dene Eskimos and Amerindian populations (11th, 12th Workshop), obtaining the genetic distances (comparison was done with about 14,660 chromosomes) (Table 1), relatedness dendrograms and correspondence analysis. In particular, the Amerindian group includes tribes from the following linguistics families: Arawakan (Wayu and Terena Indians), Chibchan (Kogi, Arhuaco, and Cayapa), Choco-Embera (Jaidukama), Ge Pano Caribe (Xavantes, Mataco and Toba), Mayan (Mayans), Mixe-Zoque (Mixe), Oto-Manguean (Mixtecans, Mazatecans and Zapotecans), Uru-Chipaya (Uros), Uto-Aztecan (Nahuas and Mayos) and Andean groups like Aymara, Quechuas and Lamas [1, 34, 35].
HLA Genotyping
Generic HLA class I (A, B and C) and high resolution HLA class II analysis (DRB1 and DQB1) was performed by PCR-SSOP-Luminex technique [62]. This methodology is fully explained in Ref. [63].
Statistical Analysis
Statistical analysis was performed with Arlequin v3.0 software [64-66].
In order to compare phenotype and haplotype HLA frequencies with other populations, the reference tables of the 11th and 12th International HLA Workshops were used [38, 67] and allelefrequencies.com web page [68]. Phylogenetic trees (dendrograms) were constructed with the allelic frequencies using the Neighbour-Joining (NJ) method [69] with the genetic distances between populations (DA) [70], using DISPAN software comprising the programs GNKDST and TREEVIEW [71, 72]. Correspondence analysis in three dimensions and its bidimensional representation was carried out using the VISTA v5.05 computer program [73], http:/forrest.psych.unc.edu).
RESULTS
Characteristic HLA Allele Frequencies Found in Wiwa Population; Comparisons with Other World Populations
The expected and observed gene frequency values for HLA-A, -B, -C, -DRB1, and -DQB1 loci do not differ significantly and the population is found in Hardy-Weinberg equilibrium (data not shown). Table 2 shows the HLA allele frequencies found in the sampled population. Twenty-three different HLA-A, twenty-eight different HLA-B, and seventeen different HLA-C alleles were found in Wiwa sample. However, only five HLA-A alleles, three HLA-B alleles, and six HLA-C alleles, (A*02:01, A*24:02, A*29:02, A*31:01, A*68:01, B*15:01, B*35:43, B*40:02, C*01:02, C*03:04, C*04:01, C*07:02, C*08:02, and C*16:01) had frequencies higher than 4%. With regard to the HLA class II alleles, nineteen different HLA-DRB1 and thirteen different HLA-DQB1 alleles were found. Only four HLA-DRB1 and four HLA-DQB1 alleles had frequencies higher than 4% (DRB1*04:07, DRB1*07:01, DRB1*08:02, DRB1*14:02, DQB1*02:02, DQB1*03:02, DQB1*04:02, and DQB1*06:02). Most alleles had already been found in Amerindians. DQB1 allele frequencies reflect the DRB1 locus allele distribution due to the strong linkage disequilibrium between these two loci.
| Allele |
Allele frequencies (%) |
Allele |
Allele frequencies (%) |
Allele |
Allele frequencies (%) |
|---|---|---|---|---|---|
| HLA-A | HLA-B | HLA-DRB1 | |||
| 01:01 | 1.8 | 35:12 | 1.8 | 01:01 | 0.9 |
| 02:01 | 10.0 | 35:14 | 0.9 | 01:02 | 0.9 |
| 02:14 | 0.9 | 35:43 | 37.3 | 03:01 | 1.8 |
| 03:01 | 3.6 | 35:44 | 1.8 | 03:02 | 0.9 |
| 11:01 | 1.8 | 35:68 | 2.7 | 04:03 | 3.6 |
| 24:02 | 42.7 | 38:01 | 1.8 | 04:05 | 2.7 |
| 24:14 | 0.9 | 39:01 | 2.7 | 04:07 | 53.6 |
| 24:17 | 0.9 | 40:02 | 5.5 | 07:01 | 6.4 |
| 24:34 | 0.9 | 40:03 | 2.7 | 08:02 | 8.2 |
| 24:57 | 0.9 | 40:09 | 1.8 | 09:01 | 0.9 |
| 26:01 | 1.8 | 40:11 | 0.9 | 11:01 | 0.9 |
| 29:02 | 4.5 | 44:03 | 3.6 | 13:01 | 1.8 |
| 30:01 | 0.9 | 44:11 | 0.9 | 13:02 | 1.8 |
| 30:02 | 1.8 | 44:45 | 0.9 | 14:02 | 7.3 |
| 31:01 | 4.5 | 51:01 | 0.9 | 14:07 | 0.9 |
| 31:12 | 3.6 | 53:01 | 0.9 | 15:01 | 3.6 |
| 33:03 | 1.8 | 15:03 | 1.8 | ||
| 33:18 | 1.8 | HLA-C | 15:33 | 0.9 | |
| 36:01 | 0.9 | 01:02 | 50.0 | 16:01 | 0.9 |
| 68:01 | 8.1 | 01:06 | 1.8 | ||
| 68:06 | 1.8 | 03:02 | 0.9 | HLA-DQB1 | |
| 68:19 | 0.9 | 03:03 | 3.6 | 02:01 | 1.8 |
| 74:01 | 0.9 | 03:04 | 8.2 | 02:02 | 7.3 |
| 03:11 | 0.9 | 03:01 | 3.6 | ||
| HLA-B | 03:28 | 0.9 | 03:02 | 62.7 | |
| 07:02 | 3.6 | 04:01 | 5.5 | 03:05 | 0.9 |
| 07:14 | 0.9 | 04:15 | 1.8 | 03:09 | 0.9 |
| 07:96 | 2.7 | 05:01 | 1.8 | 03:18 | 2.7 |
| 14:01 | 1.8 | 06:14 | 0.9 | 04:02 | 8.2 |
| 14:02 | 0.9 | 07:02 | 8.2 | 05:01 | 2.7 |
| 14:11 | 1.8 | 07:05 | 0.9 | 05:02 | 0.9 |
| 15:01 | 12.7 | 08:02 | 4.5 | 06:02 | 5.5 |
| 18:01 | 1.8 | 12:03 | 2.7 | 06:03 | 1.8 |
| 24:02 | 0.9 | 15:02 | 0.9 | 06:09 | 0.9 |
| 35:01 | 0.9 | 16:01 | 4.5 | ||
| 35:04 | 0.9 | ||||
| 35:05 | 1.8 |
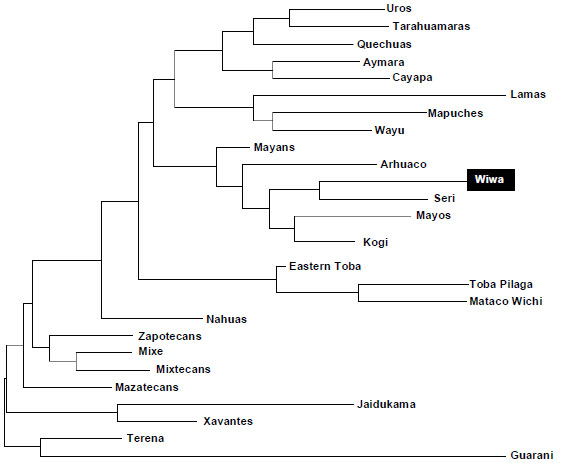
Two types of analysis were done in order to compare Wiwa HLA frequencies with other World population frequencies: 1) with pooled DRB1 and DQB1 data (data not shown); and 2) with DRB1 only. It was not possible to carry out a study comparing HLA class I allele frequencies or HLA class I and II conjointly due to the lack of class I studies in many World wide populations. A single DRB1 study was carried out in order to compare the Wiwa HLA population frequencies with those of Caucasoids and Siberians who lacked DQB1 analyses. The Neighbour-Joining (NJ) relatedness dendrogram based on HLA-DRB1 allele frequencies (Fig. 2) separates all the populations in two well-differentiated clusters, one of them shows Amerindian grouped together with North American Na-Dene and Eskimo, as well as Siberians. The other cluster grouped the rest of World wide populations. Wiwa are integrated in the Amerindian group, close to Mexican Mayans. Bootstrap values were 100% in all nodes.
The correspondence analysis based on HLA-DRB1 allele frequencies (Fig. 3) and on HLA-DRB1 and HLA-DQB1 frequencies conjointly (data not shown) give similar results: Amerindians (including Wiwa) form a single and compact group separated from the rest of World wide populations, which show a continuous genetic variation from Siberia-East Asia to the Mediterranean (Fig. 3). The North American Na-Dene and Eskimo seem to be closer to Siberian and Orientals than to Amerindians. Lastly, a correspondence analysis based on HLA-DRB1 allele frequencies and on HLA-DRB1 and HLA-DQB1 frequencies conjointly (data not shown) including only Amerindian populations was done to study the possible correlation between genes and languages, and it is observed that populations genetically close speak languages belonging to different and distant linguistic families [34].

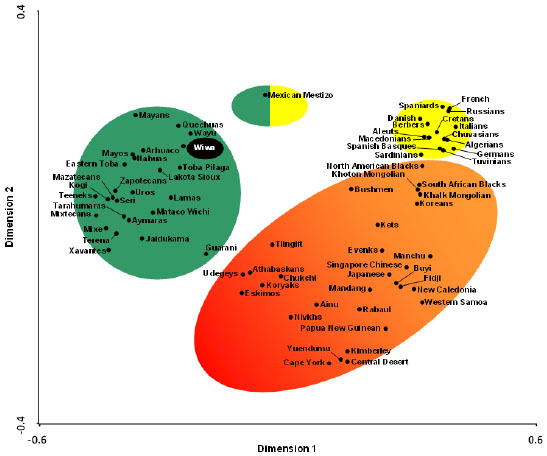
Genetic distances between Wiwa and other populations show that Wiwa are both close to Meso and South Americans (Table 3). Indeed, Mayans (Guatemala, Mayan linguistic family), Seri (Mexico, according to some authors belonging to Hokan linguistic family, according to others is a language isolate) [74], Kogi (Colombia, Chibchan linguistic family), Arhuaco (Colombia, Chibchan linguistic family) and Mayos (Mexico, Uto-Aztecan linguistic family) show the closest genetic distances to Wiwa, followed by Mexican Mestizo, Tarahumara (Mexico, Uto-Aztecan linguistic family), Quechuas (Bolivia, Quechuan linguistic family), Uros (Peru, Uru-Chipaya linguistic family) and the rest of Amerindian populations. A genetic distance values discontinuity between Amerindians and the other world groups (including Na-Dene and Eskimos) is observed. With respect to Guarani population, we could observe that a higher genetic distance exists with Wiwa; larger than with other non Amerindian population (Athabaskans), this fact may be due to the low number of HLA typings of the Guarani population or to admixture (Table 3). Again, these results show that genes and languages do not seem to correlate at the microgeographical level [1, 34].
| Population | DA (%) | Population | DA (%) |
|---|---|---|---|
| Mayans | 13.41 | Japanese | 59.09 |
| Seri | 17.81 | Algerians | 59.53 |
| Kogi | 18.15 | Albanians | 59.62 |
| Arhuaco | 22.43 | South African Blacks | 59.99 |
| Mayos | 22.98 | Spanish Basques | 60.25 |
| Mexican-Mestizo | 25.57 | Ashkenazi Jews | 60.27 |
| Tarahumara | 27.93 | Non-Ashkenazi Jews | 60.43 |
| Quechuas | 28.58 | Cretans | 60.54 |
| Uros | 30.01 | Mataco-Wichi | 60.75 |
| Sioux-Lakota | 32.11 | Aleuts | 61.58 |
| Mixtecans | 32.85 | Lebanese KZ | 61.87 |
| Cayapa | 33.28 | Moroccan Jews | 63.36 |
| Teenek | 33.76 | Manchu | 63.48 |
| Nahuas | 35.04 | Sardinians | 63.65 |
| Wayu | 35.05 | Macedonians | 63.91 |
| Aymaras | 36.32 | Udegeys | 64.36 |
| Lamas | 38.71 | Chukchi | 66.43 |
| Mazatecans | 38.95 | Western Samoa | 66.61 |
| Mapuches | 39.76 | Nivkhs | 66.75 |
| Zapotecans | 40.78 | Khoton Mongolian | 68.89 |
| Eastern Toba | 42.27 | Koryaks | 68.93 |
| Mixe | 43.55 | Eskimos | 69.40 |
| Jaidukama | 43.65 | Singapore Chinese | 70.26 |
| Toba-Pilaga | 43.74 | Bushmen | 71.87 |
| Athabascans | 47.14 | New Caledonia | 71.99 |
| Tuvinians | 48.12 | Lebanese NS | 72.15 |
| Xavantes | 50.43 | Ainu | 72.76 |
| Spaniards | 52.15 | Fidji | 74.43 |
| Germans | 52.77 | Buyi | 75.47 |
| French | 53.49 | Kets | 79.54 |
| Koreans | 53.62 | Evenks | 79.87 |
| North American Blacks | 54.24 | Papua New Guinea | 81.63 |
| Tlinglit | 54.27 | Guarani | 82.21 |
| Russians | 54.93 | Rabaul | 83.43 |
| Terena | 55.87 | Madang | 83.79 |
| Berbers (Souss) | 57.03 | Cape York | 86.55 |
| Khalk Mongolian | 57.11 | Central Desert | 89.75 |
| Italians | 57.52 | Yuendumu | 94.36 |
| Chuvashians | 57.75 | Kimberley | 96.58 |
| Danish | 57.93 |
| Haplotype (A-B-C-DRB1-DQB1) | HF (%) | Possible origin |
|---|---|---|
| A*24:02-B*35:43-C*01:02-DRB1*04:07-DQB1*03:02a | 18.2 | Amerindian |
| A*02:01-B*35:43-C*01:02-DRB1*04:07-DQB1*03:02b | 5.5 | Amerindian |
| A*24:02-B*15:01-C*01:02-DRB1*04:07-DQB1*03:02c | 4.5 | Amerindian |
| A*29:02-B*44:03-C*16:01-DRB1*07:01-DQB1*02:02d | 3.6 | Western Europe |
| A*31:01-B*35:43-C*01:02-DRB1*04:07-DQB1*03:02e | 3.6 | Amerindian |
| A*68:01-B*15:01-C*03:03-DRB1*14:02-DQB1*03:02f | 2.7 | New |
| A*11:01-B*07:02-C*07:02-DRB1*15:03-DQB1*06:02g | 1.8 | New |
| A*24:02-B*07:02-C*07:02-DRB1*15:01-DQB1*06:02h | 1.8 | Eastern Europe |
| A*24:02-B*35:44-C*01:02-DRB1*04:07-DQB1*03:02i | 1.8 | Amerindian |
| A*24:02-B*40:03-C*01:02-DRB1*04:07-DQB1*03:02j | 1.8 | Amerindian |
| A*26:01-B*38:01-C*12:03-DRB1*13:01-DQB1*06:03k | 1.8 | Western Europe |
| A*68:01-B*15:01-C*03:04-DRB1*14:02-DQB1*03:01l | 1.8 | New |
a Found without HLA-C in Mayans (5%), Teeneks (3.7%), Seri (2.3%), Lakota-Sioux (2.2%), Aymaras (2.1%) and Quechuas (1.4%).
b Found without HLA-C in Seri (18.2%), Teeneks (15.5%), Mayans (10.6%), Mixtecans (3%), Mazatecans (2.5%), Aymaras (1.7%) and Peruvian Indians (1.7%).
c Found without HLA-C in Mayans (2.3%).
d Found in Irish (1.3%), Tunisians (4.0%) and without HLA-C in Murcians (5.1%) and in Amerindians immigrants to Madrid (0.8%).
e Found without HLA-C in Mayans (2.6%) and Quechuas (1.4%).
f, g, l Not found in any other population.
h Found without HLA-C in Chuvash (4.3%).
i Found without HLA-C in Mayans (5%), Teeneks (3.7%), Seri (2.3%), Lakota-Sioux (2.2%), Aymaras (2.1%) and Quechuas (1.4%).
j Found without HLA-C in Mayos (1.7%).
k Found without HLA-C in Murcians (1.9%).
References: [16, 26-28, 31, 52, 53, 75-78].
HLA-A, -B, -C, -DRB1 and -DQB1 Extended Haplotype Analysis in Wiwa: Comparison with Other Populations
Associations between different HLA loci were estimated. The most probable two loci haplotype combinations (A-B, B-DRB1 and DRB1-DQB1) were calculated (data not shown). The twelve most frequent five HLA loci haplotype combinations (A-B-C-DRB1-DQB1) were calculated and these extended haplotypes are depicted in Table 4; they represent 48.9% of all haplotypes. Wiwa extended HLA haplotypes have been obtained for the first time, allowing their comparison with previously reported ones in other populations (Table 4 and its footnote). The partial class II haplotype DRB1*04:07-DQB1*03:02 is present in association with A*24:02-B*35:43-C*01:02, A*02:01-B*35:43-C*01:02, A*24:02-B*15:01-C*01:02, A*31:01-B*35:43-C*01:02, A*24:02-B*35:44-C*01:02, and A*24:02-B*40:03-C*01:02 (35.4% of total haplotypes). The majority of most frequent extended HLA haplotypes have Amerindian characteristics and they have been found in other Amerindian populations, mainly in Meso-American populations (Mayans, Mazatecans, Mixtecans, Seri, Teeneks) [26, 29, 53] and South-American populations (Aymaras and Quechuas) (Table 4) [16, 31]. Haplotypes (or partial haplotypes) with no Amerindian characteristics have been found in this sample: one of them has been found in Chuvash population in high frequency (4.3%) [52] and two extended haplotypes are present in Murcian population [75]. The rest of extended HLA haplotypes (6.3%) have not been found in any other World wide ethnic group, these haplotypes seem to be specific for this Wiwa-Colombian ethnic group.
In summary, most extended HLA haplotypes in our populations sample are either from Amerindian origin or newly found and bearing typical HLA Amerindian alleles.
High Frequency Wiwa HLA alleles: Their Comparison with Pacific and Other World Populations
Due to the high frequency of HLA-A*24:02 and HLA-C*01:02 alleles in our study population, (see Table 2) a World distribution map of these alleles was performed (see Fig. 4). That map shows that the highest frequencies of these alleles are found in the Pacific area. Significant differences on these two alleles frequency values were found between Pacific populations and Europe Africa, continental and West Asia populations, based on t-student test (data not shown).
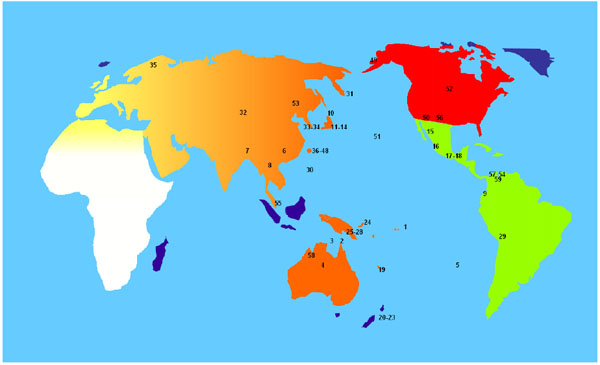
1- American Samoa: A*24:02→33.00%; C*01:02→24.00%.
2- Australia Cape Cork Peninsula Aborigine: A*24:02→22.30% [79].
3- Australia Groote Eylandt Aborigine: A*24:02→29.30%; C*01:02→26.70% [79].
4- Australia Yuendumu Aborigine: A*24:02→29.80; C*01:02→24.70% [79].
5- Chile Easter Island; A*24:02→35.80%; C*01:02→21.40% [80].
6- China Guizhou Province Shui: A*24:02→24.30%; C*01:02→28.40% [81].
7- China Tibet Region Tibetan: A*24:02→27.20% [82].
8- China Yunnan Bulang: A*24:02→23.70% [83].
9- Ecuador Cayapa: A*24:02→61.40% [43].
10- Japan Hokkaido Ainu: A*24:02→24.00% [60].
11- Japan Central: A*24:02→37.90% [84].
12- Japan pop 16: A*24:02→36.48% [85].
13- Japan pop 3: A*24:02→36.20% [62].
14- Japan pop5: A*24:02→32.70% [86].
15- Mexico Chihuahua Tarahumara: A*24:02→37.50% [32].
16- Mexico Mixtec: A*24:02→25.30% [87].
17- Mexico Oaxaca Mixtec: A*24:02→24.50% [88].
18- Mexico Oaxaca Zapotec: A*24:02→23.10% [88].
19- New Caledonia: A*24:02→60.70%; C*01:02→37.80% [89].
20- New Zealand Maori with admixed history: A*24:02→29.10%; C*01:02→31.40% [90].
21- New Zealand Maori with full ancestry: A*24:02→38.00%; C*01:02→44.60% [90].
22- New Zealand Polynesians with admixed history: A*24:02→33.30%; C*01:02→27.80% [90].
23- New Zealand Polynesians with full ancestry: A*24:02→31.00%; C*01:02→21.40% [90].
24- Papua New Guinea East New Britain Rabaul: A*24:02→31.60% [89].
25- Papua New Guinea Eastern Highlands Goroka Asaro: A*24:02→74.40%; C*01:02→30.20% [89].
26- Papua New Guinea Karimui Plateau Pawaia: A*24:02→74.40%; C*01:02→20.00% [89].
27- Papua New Guinea Waginela Keapara: A*24:02→62.70% [89].
28- Papua New Guinea Wosera Abelam: A*24:02→51.30% [89].
29- Peru Titikaka Lake Uro: A*24:02→28.10% [14].
30- Philippines Ivatan: A*24:02→32.00%. [91].
31- Russia Bering Island Aleut: A*24:02→24.10% [92].
32- Russia Tuva pop 2: A*24:02→21.50% [93].
33- South Korea pop 10: A*24:02→22.09% [94].
34- South Korea pop 3: A*24:02→21.70% [95].
35- Sweden Northern Sami: A*24:02→21.20% [96].
36- Taiwan Ami: A*24:02→62.80%; C*01:02→21.90% [91].
37- Taiwan Atabal: A*24:02→61.80% [91].
38- Taiwan Bunun: A*24:02→58.40% [91].
39- Taiwan Paiwan: A*24:02→86.30% [91].
40- Taiwan Pazeh: A*24:02→33.60% [91].
41- Taiwan Puyuma: A*24:02→ 64.00% [91].
42- Taiwan Rukai: A*24:02→76.00% [91].
43- Taiwan Saisiat: A*24:02→59.90% [91].
44- Taiwan Siraya: A*24:02→47.10% [91].
45- Taiwan Tao: A*24:02→54.00% [91].
46- Taiwan Taroko: A*24:02→44.50% [91].
47- Taiwan Thao: A*24:02→60.00% [91].
48- Taiwan Tsou: A*24:02→78.40% [91].
49- USA Alaska Yupik: A*24:02→58.10% [97].
50- USA Arizona Gila River Amerindian: A*24:02→34.10% [98].
51- USA Hawai Okinawa: A*24:02→34.30%; C*01:02→20.50%.
52- USA South Dakota Lakota Sioux: A*24:02→26.20% [28].
53- Mongolia Buryat: A*24:02→20.00%.
54- Venezuela Sierra de Perija Yucpa: A*24:02→23.30 [99].
55- Singapore Chinese Han: A*24:02→21.50%.
56- USA New Mexico Canoncito Navajo: A*24:02→30.50%.
57- Venezuela Perja Mountain Bari: A*24:02→60.20%; C*01:02→21.20%.
58- Australia Kimberly Aborigine: C*01:02→37.50% [79].
59- Wiwa: A*24:02→42.70%; C*01:02→50.00% (present study).
HLA Frequencies Between Wiwa and Other Amerindians
The comparison between Wiwa and Amerindians HLA frequencies were performed in order to determine the relationship grade inside this group of populations. In Fig. (5) can be observed a Neighbour-Joining (NJ) dendrogram based on HLA-DRB1 allele frequencies that shows this relatedness. The bootstrap values of each of the nodes were 100%.
DISCUSSION
Wiwa Amerindian Population and America Peopling According to HLA Genes
It is clear that Wiwa-Colombians HLA analysis supports our previous conclusions about peopling of America and relationship with other groups:
- Amerindians tend to group together and separate from other World populations in our statistical analyses (Table 3 and Fig. (3), and also in Fig. (2) at a certain extent) [26, 29, 33, 100].
- Wiwa Chibchan language [1] does not mean that they are genetically closer to other Chibchan language speaking groups (Table 3, Figs. 2, 3 and 5). In some analyses they are closer to Mayans (Mayan language, Fig. 2) and to Seri Fig. (2). Also, they go close to Wayu (Arawak speaking group) [1] in correspondence analyses Fig. (3).
- Additional HLA frequencies analyses in this paper, which include Wiwa Amerindians, do not support a three waves model for America peopling [29, 100]. This also supports analyses done with other genetic markers (mtDNA, YChr, or Alu sequences [3, 4, 9, 10].
In addition, a multidisciplinary approach is necessary because only genetics is not enough to accurately approach to America peopling [100]. It is evident that an ancient founder effect may have disappeared because of a continuous new population admixture effect. Also, European carried diseases caused that about 85% of America First Inhabitants died after 1492 (AD) because of newly introduced microbes [101]. This may have changed HLA profile of Amerindians after 1492 (AD). Stress (like epidemics) causes appearance of new HLA alleles in spermatozoa as have beeen shown by single spermatozoa PCR [102]. A set of new alleles may have appeared in Amerindians after 1492.
Moreover, our present analyses are useful for implementing programs of HLA pharmacogenomics [12, 87, 103] and preventive medicine of diseases linked to HLA in Wiwa and other Amerindians. Transplantation programs will also benefit when established in Amerindian areas [12, 87, 103].
HLA Extended Haplotypes in Wiwa and Geographical Distribution of High Frequency A*24:02 and C*01:02 Alleles
This is the first time that HLA-A, -B, -C, -DRB1 and DQB1 extended haplotypes have been calculated in Wiwa. A confirmation that Wiwa group has a low admixture degree is that A*24:02-B*35:43-C*01:02-DRB1*04:07-DQB1*03:02 most frequent extended haplotype has a very high frequency (18.2%, compared to the second most frequent haplotype A*02:01-B*35:43-C*01:02-DRB1*04:07-DQB1*03:02). Most haplotypes are of Amerindian origin, since they have already been described in Table 4.
Three new extended haplotypes have been found: A*68:01-B*15:01-C*03:03-DRB1*14:02-DQB1*03:02; A*11:01-B*07:02-C*07:02-DRB1*15:03-DQB1*06:02 and A*68:01-B*15:01-C*03:04-DRB1*14:02-DQB1*03:01, which are characteristic of this particular population.
On the other hand, striking A*24:02 and C*01:02 high frequencies were found and also these alleles are present in the most frequent Wiwa HLA extended haplotypes.
Taken into account our previous detection of HLA alleles and haplotypes shared by Amerindians and Pacific Islanders [12, 103], we undertook a frequency comparison of A*24:02 and C*01:02 alleles with Pacific populations, which are also frequent in other Amerindians (Fig. 4) [12, 103, 104].
These alleles were certainly found with a high frequency (compared to European populations) in some Pacific Ocean populations:
- A*24:02: Koreans (22.09%); Taiwanese isolates (86.3%, being the maximum frequency); Papua New Guinea (74.4%), New Caledonia (60.70%, one of the highest A*24:02 frequencies); New Zealand (38%); Japan (37.90%). Amerindians also presented A*24:02 high frequencies (see Fig. 4).
- C*01:02: New Zealand (44.60%); New Caledonia (37.80%); Australia Kimberley Aborigine (37.50%); Papua New Guinea (30.20%); Taiwan (21.90%).
These Pacific populations A*24:02 and C*01:02 frequencies are statistical significantly higher when compared to Europeans, Africans and continental and West Asia populations (data not shown).
Altogether [12, 103] our findings in this genealogical A*24:02, C*01:02 alleles study show that HLA support that Pacific Islanders and Amerindians have had genetic exchange [12, 103] in ancient times. Cultural and other partial genetic traits also show that Amerindians have had genetics contacts with Pacific Easter Island [16, 17, 80]. Also, a widespread SNPs analyses cast doubts about Amerindian genes found admixed with Eurasian before 1492 (Columbus) but only three clear Amerindian populations are analysed [105].
Our conclusion on Amerindians and Pacific Islands relatedness is that genetic and cultural common traits [12, 103] may have been shared through a both directions traffic.
Finally, there are cultural [25] and genetic evidences (mtDNA analyses) [106-109] that show that Solutrean Europeans shared them with Ancient American inhabitants [106-109]. Therefore, simple models of America population should be revised.
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported in part by Grants from the Spanish Ministry of Health and Economy (PI11/00807 and PI14/01067), European FEDER funds and three different Mutua Madrileña Automovilista grants.


