All published articles of this journal are available on ScienceDirect.
Kurds HLA Genes: Its Implications in Transplantation and Pharmacogenomics
Abstract
HLA genes (class I and II) have been studied in a Kurd population from Iran (North West towns of Saqqez and Baneh, close to Irak border). Kurds speak an Iranian language. HLA Kurd profile has been compared with those of Central Asians, Siberians, Mediterraneans and other worldwide populations; a total of 7746 chromosomes were used for computer comparisons. Both Neighbor-joining and correspondence genetic analyses place Kurds in the Mediterranean population cluster, close to Iranians, Europeans and Caucasus populations (Svan and Georgian). New extended HLA haplotypes are described, being A*02:01-B*35:01-DRB1*01:01-DQB1*05:01 and A*24:02-B*18:09-DRB1*11:01- DQB1*03:01 the most frequent ones; other Kurd extended haplotypes are also found in Azeris and Palestinians. This research work may be useful for: 1) future Iranian Kurds transplantation regional programs, 2) HLA pharmacogenomics in order to practise a preventive Medicine and drug side effects, and 3) Epidemiology of HLA-associated diseases in Kurds.
INTRODUCTION
The HLA system is the most polymorphic genetic system described in humans. It consists of several closely linked loci encoding cell surface glycoproteins whose main function is activating immune system response through antigenic presentation. New loci and new alleles have continuously been described since HLA discovery (i.e.:1582 HLA-DRB1 alleles have been described up to October 2014; [1]). HLA gene frequencies have both a large degree of variability, and a remarkable geographical correlation and they are a very useful tool to infer genetic background and ethnical composition of modern human populations and also for tracing migration of ancient ones; e.g.: certain alleles are frequent only in specific populations (i.e. A36:01, A43:01 in Negroids [2]). In addition, certain combinations of contiguous alleles due to the strong linkage disequilibrium between HLA neighbouring loci show a characteristic frequency or are distinctive in many present day populations [3]. Also, HLA genetic system is a unique tool for studying the origins of relatively isolated groups, like the Kurd people living in Iran (see below, Fig. 1). Finally, HLA alleles are linked to diseases and also to response to drug treatments, which accomplishes relationships of certain HLA alleles with at least fifteen different pathologies treatment including AIDS [4]. This is important in personalized treatments design.

Map of the area referred in the text. Green dots area defines Kurds distribution (Kurdistan); small dots: high density; big dots: low density. Black dots represent location of study samples.
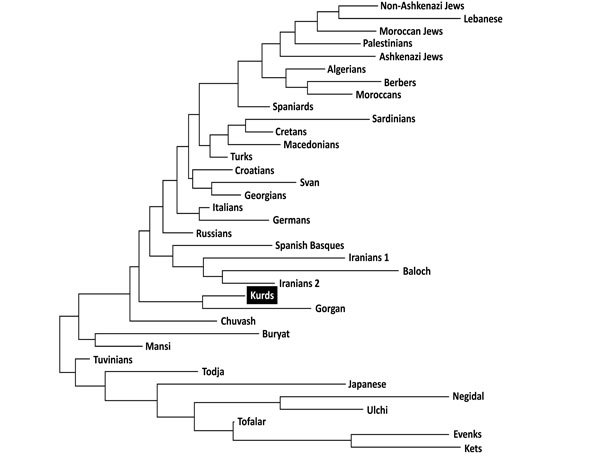
Neighbor-Joining (NJ) dendrogram showing relatedness between Kurds and other World populations. Genetic distances between populations (DA) were calculated by using HLA-DRB1 (high resolution). Data from World populations were taken from references stated in Table 1.
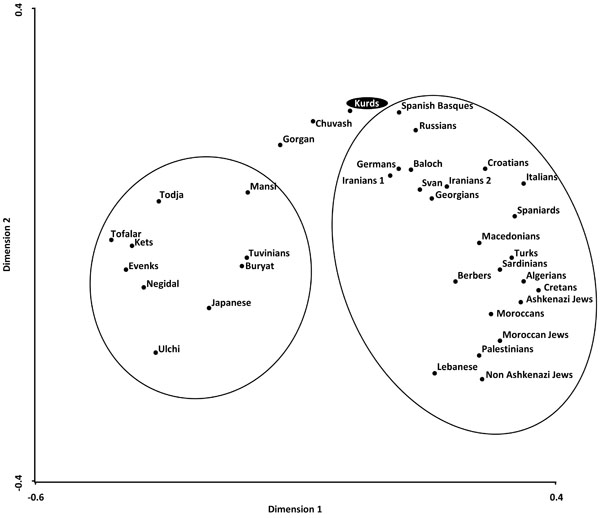
Correspondence analysis showing a global view of the relationship between Kurds and Mediterranean, Siberians, and other World populations according to HLA-DRB1 allele frequencies in three dimensions (bidimensional representation).
Worldwide populations included in the analysis. A total of 7746 chromosomes were analysed. N, number of individuals.
| Population | N | Reference | Population | N | Reference |
|---|---|---|---|---|---|
| Algerians | 51 | [40] | Lebanese | 59 | [22] |
| Ashkenazi Jews | 80 | [41] | Macedonians | 178 | [45] |
| Baloch | 100 | [42] | Mansi | 68 | [30] |
| Berbers (Souss) | 98 | [33] | Moroccans | 98 | [46] |
| Buryat | 25 | [30] | Moroccan Jews | 113 | [47] |
| Chuvash | 82 | [29] | Negidal | 35 | [30] |
| Cretans | 144 | [43] | Non Ashkenazi Jews | 80 | [41] |
| Croatians | 105 | [22] | Palestinians | 165 | [48] |
| Evenks | 35 | [31] | Russians | 200 | [49] |
| Germans | 295 | [21] | Sardinians | 91 | [21] |
| Georgians | 119 | [44] | Spaniards | 88 | [50] |
| Gorgan | 69 | [28] | Spanish Basques | 83 | [50] |
| Iranians 1a | 65 | [1] | Svan | 80 | [51] |
| Iranians 2b | 73 | [1] | Turks | 245 | [52] |
| Italians | 284 | [21] | Todja | 22 | [30] |
| Japanese | 493 | [21] | Tofalar | 43 | [30] |
| Kets | 22 | [31] | Tuvinians | 197 | [3] |
| Kurds | 60 | This study | Ulchi | 73 | [30] |
a Iranians from Yazd province.
b Iranians from Fars province.
HLA-A, -B, -DRB1, and -DQB1 allele frequencies in Kurds population.
| Alleles | Allele Frequencies % | Alleles | Allele Frequencies % | Alleles | Allele Frequencies % |
|---|---|---|---|---|---|
| HLA-A | 15:17 | 0.8 | 03:01 | 15.8 | |
| 01:01 | 8.3 | 15:24 | 0.8 | 04:01 | 12.5 |
| 01:02 | 2.5 | 18:01 | 5.8 | 04:15 | 0.8 |
| 02:01 | 12.5 | 18:09 | 3.3 | 07:01 | 6.7 |
| 02:48 | 1.7 | 27:01 | 0.8 | 08:01 | 1.7 |
| 02:50 | 5.8 | 35:01 | 22.0 | 10:01 | 1.7 |
| 02:52 | 6.7 | 35:20 | 0.8 | 11:01 | 19.1 |
| 02:61 | 1.7 | 35:27 | 1.7 | 12:01 | 0.8 |
| 03:01 | 10.8 | 38:01 | 4.2 | 13:01 | 5.0 |
| 03:02 | 0.8 | 40:01 | 1.7 | 13:02 | 4.2 |
| 11:01 | 8.3 | 40:15 | 0.8 | 13:05 | 0.8 |
| 13:01 | 0.8 | 41:01 | 0.8 | 14:01 | 6.7 |
| 23:01 | 2.5 | 44:02 | 5.0 | 15:01 | 10.0 |
| 24:02 | 17.6 | 44:10 | 0.8 | 16:01 | 4.2 |
| 24:03 | 0.8 | 49:01 | 5.0 | HLA-DQB1 | |
| 26:01 | 7.5 | 50:01 | 5.8 | ||
| 30:01 | 1.7 | 51:01 | 9.2 | 02:01 | 20.8 |
| 31:01 | 1.7 | 51:06 | 0.8 | 02:02 | 0.8 |
| 32:01 | 5.0 | 51:07 | 0.8 | 03:01 | 24.2 |
| 33:01 | 1.7 | 52:01 | 4.2 | 03:02 | 6.7 |
| 68:01 | 0.8 | 53:01 | 2.5 | 03:05 | 4.2 |
| 69:01 | 0.8 | 54:01 | 0.8 | 03:07 | 1.7 |
| HLA-B | 55:01 | 5.0 | 04:01 | 2.5 | |
| 58:01 | 5.0 | 04:02 | 0.8 | ||
| HLA-DRB1 | 05:01 | 10.8 | |||
| 07:02 | 1.7 | 05:02 | 7.5 | ||
| 07:24 | 0.8 | 06:01 | 5.0 | ||
| 08:01 | 5.0 | 01:01 | 7.5 | 06:02 | 6.7 |
| 13:01 | 3.3 | 01:02 | 0.8 | 06:03 | 5.0 |
| 14:01 | 0.8 | 01:03 | 1.7 | 06:04 | 3.3 |
Genetic distances (DA) between Kurds and other populations (x10-2) obtained by using HLA-DRB1 allele frequencies.
| Population | DA |
|---|---|
| Gorgan | 13.41 |
| Russians | 14.86 |
| Italians | 15.73 |
| Croatians | 16.09 |
| Georgians | 16.26 |
| Svan | 17.26 |
| Germans | 19.57 |
| Spanish Basques | 19.63 |
| Iranians 2 | 20.12 |
| Turks | 20.48 |
| Chuvash | 20.92 |
| Macedonians | 21.87 |
| Spaniards | 22.04 |
| Tuvinians | 23.96 |
| Cretans | 24.73 |
| Berbers | 25.56 |
| Todja | 25.71 |
| Mansi | 27.29 |
| Tofalar | 29.65 |
| Baloch | 29.85 |
| Sardinians | 30.06 |
| Algerians | 31.12 |
| Iranians 1 | 31.37 |
| Palestinians | 33.47 |
| Moroccans | 33.49 |
| Ashkenazi Jews | 34.57 |
| Moroccan Jews | 35.38 |
| Buryat | 36.55 |
| Non Ashkenazi Jews | 37.06 |
| Lebanese | 39.78 |
| Ulchi | 47.76 |
| Evenks | 47.90 |
| Negidal | 52.61 |
| Kets | 53.07 |
| Japanese | 53.22 |
The twelve most frequent HLA-A, -B, -DRB1 and -DQB1 extended haplotypes in Kurds.
| Haplotypes | HF (%) | Possible Origin |
|---|---|---|
| A*02:01-B*35:01-DRB1*01:01-DQB1*05:01a | 3.3 | Mediterraneans and others |
| A*24:02-B*18:09-DRB1*11:01-DQB1*03:01b | 3.3 | New |
| A*01:01-B*35:01-DRB1*11:01-DQB1*03:01c | 2.5 | Mediterranean |
| A*03:01-B*44:02-DRB1*11:01-DQB1*03:01d | 2.5 | Mediterraneans and others |
| A*24:02-B*52:01-DRB1*15:01-DQB1*06:01e | 2.5 | New |
| A*24:02-B*58:01-DRB1*03:01-DQB1*02:01f | 2.5 | Mediterraneans, Orientals and others |
| A*01:01-B*08:01-DRB1*03:01-DQB1*02:01g | 1.7 | Mediterranean |
| A*02:01-B*35:01-DRB1*11:01-DQB1*03:01h | 1.7 | (Azeris) |
| A*02:01-B*50:01-DRB1*07:01-DQB1*02:01i | 1.7 | Caucasian-Mediterranean |
| A*11:01-B*50:01-DRB1*07:01-DQB1*02:01j | 1.7 | Mediterraneans, Orientals and possibly Amerindians |
| A*24:02-B*51:01-DRB1*08:01-DQB1*04:01k | 1.7 | New |
| A*26:01-B*08:01-DRB1*03:01-DQB1*02:01l | 1.7 | Mediterraneans, Orientals and others |
HF: haplotipic frequency.
aFound in different frequencies in Caucasoids, Africans and possibly Amerindians.
b,e,kNot found in any other population
dFound in different frequencies in Caucasoids and Africans.
cFound with HLA-Class I in low resolution in Palestinians (HF: 1.2)
fFound in different frequencies in Caucasoids and Orientals.
g Found in South Ireland (HF: 11.5) and Northwest England (HF: 9.5) and with HLA-class I in low resolution in Chuvash (HF: 1.2)
hFound in Azeris (HF: 1.1, unpublished data)
lFound in differents frequencies in Caucasoids, Africans and Orientals.
jFound in differents frequencies in Caucasoids, Orientals and possibly in AmerindiansiFound in Azeris (HF: 1.1, unpublished data) and in Armenians (HF: 1.0)
References: [1, 29, 48]
HF: haplotipic frequency
aHLA class I split found in population of Oman (HF: 4.9). HLA Class II split found in Chuvash (HF: 18.7), Sami (HF: 9.2), Tuvinians (HF: 9.0), Khoton Mongolian (HF: 8.1), Mansi (HF: 6.0) and Aleuts (HF: 5.6)
b,d HLA class II split found in Sami (HF: 11.7), Turkish (HF: 9.8), Tofalar (HF: 9.3), Todja (HF: 9.1), Kets (HF: 8.8), Evenks (HF: 8.6) and Chuvash (HF: 4.8)
cFound with HLA-class I in low resolution in Palestinians (HF: 1.2)
eHLA class II split found in Indian Marathas (HF: 22.9), Kets (HF: 2.9) and Khoton Mongolian (HF: 2.4)
fHLA class II split found in Sardinians (HF: 25.3), Souss Berbers (HF: 17.3), Khoton Mongolian (HF: 13.6) Iranians from Fars (HF: 11.0) and Mansi (HF: 7.4)
g Found in South Ireland (HF: 11.5) and Northwest England (HF: 9.5) and with HLA-Class I in low resolution in Chuvash (HF: 1.2)
hFound in Azeris (HF: 1.1, unpublished data)
iFound in Azeris (HF: 1.1, unpublished data) and in Armenians (HF: 1.0)
jHLA class I split found in Moroccans (HF: 3.6). HLA class II split found in Buryat (HF: 22.0), Berbers (HF: 20.1), Mansi (HF: 16.9), Iranians from Yazd (HF: 15.4) and Kets (HF: 11.8)
kNot found in any other population
lNot found in any other population
References: [1, 3, 29, 30, 31, 32, 33, 48, 52, 53]
HLA genetic studies in Turkish and Kurdish populations suggested a common Anatolian-Mediterranean origin in both populations; Kurds, probably coming from old Hurrians [5, 6]. Genetic studies based on mitochondrial DNA (mtDNA) and Y chromosome (Y-chr), have been also carried out on Kurds, but there is not much concordance between these two genetic markers to infer the Kurd people origins that may probably be from Middle East and/or Central Asia [7, 8].
Kurdistan (“land of Kurds”) is a geographical and cultural region located South of Caucasus and North of the former Mesopotamia. Present day Kurd gene pool may be due to admixture of ancient North Mesopotamian and Central Asia populations, who share common culture origins [9-13].
Kurds live nowadays in Turkey, Syria, Iraq, Armenia, Turkmenistan, Kazakhstan and Iran (Fig. 1). They are mainly defined by language speaking and are estimated to be about 30 to 38 million people [14, 15]. In Iran, Kurds live mainly in Iranian Kurdistan (Northeast part of the Country) and amount about 5 million people. A Kurd community there exists in East Iran, Khorasan, near Turkmenistan boundary [16] (Fig. 1). Nowadays in Iran, they form both urban and rural communities, but there are also a few groups that still practice nomadism [16].
We have undertaken the present study in order to: 1) Determine the HLA class I (A and B) and class II (DRB1 and DQB1) allelic Kurd lineages (hereafter ‘‘alleles’’ for simplicity) and specific HLA haplotypes by using PCR-SSOP-Luminex and DNA sequencing, 2) Compare the Kurd HLA allele frequencies with those Central Asian, Siberian, Mediterranean and other worldwide populations (Table 1) with computer programs in order to establish bases of HLA disease epidemiology and origins in Kurds by constructing groups of genes frequencies comparisons using genetic distances, Neighbour Joining (NJ) dendrograms and correspondence analyses and finally, 3) Establishing the Kurd HLA profile that will also be useful for preventive HLA pharmacogenomics and a virtual regional future transplant waiting list among populations.
MATERIAL AND METHODS
Population Sample
60 healthy unrelated volunteers from the cities of Saqqez and Baneh, Iran (Fig. 1) were HLA class I and class II typed. The cities of Saqqez and Baneh are located in Kurdistan province in the North East of Iran (36°14′47″N 46°15′59″E and 35°59′51″N 45°53′07″E respectively, see Map, Fig. 1) and both are historically and continuously inhabited by Kurds since 1000 BC. A written consent to participate in the present study was signed by each individual. All subjects in the study were born in these two cities and their four grandparents had been born in the same area [16]. We compare our data with those of Caucasian European, Mediterranean, Siberian and Oriental populations (these populations are detailed in Table 1), obtaining the genetic distances (comparison was done with 7,746 chromosomes), relatedness dendrograms and correspondence analyses.
HLA Genotyping
Generic HLA class I (A and B) and high resolution HLA class II analysis (DRB1 and DQB1) was performed by PCR-SSOP-Luminex technique [17]. This methodology consists of: a) PCR using specific primer pairs as provided by manufacturers (Luminex Corporation, Austin, TX, USA). All of these primers are 5’-biotined and they are specific to determinate sequences of exons 2 and 3 (or only exon 2 for HLA class II) of HLA genes; b) hybridization: products of PCR biotin-labelled were denaturalized at 97 ºC and then were hybridized to complementary DNA probes associated to microbeads; and c) assignation of HLA alleles: complex resulting of hybridization was introduced a Luminex platform, this system identify the fluorescent intensity of fluorophores on each oligobead that has hybridized with the biotin-labelled PCR product. Luminex Software assigns the HLA alleles for each DNA sample [17]. HLA-A, -B, -DRB1, and -DQB1 allele DNA automated sequencing (ABI PRISM 3700/ ABI PRISM 3730. Applied Biosystems; California) was only done when DNA typing yielded ambiguous results.
Statistical Analysis
Statistical analysis was performed with Arlequin v2.0 software kindly provided by Excoffier and Slatkin [18]. In summary, this program calculated HLA-A, -B, -DRB1, and -DQB1 allele frequencies, Hardy-Weinberg equilibrium and the linkage disequilibrium between n alleles at n different loci. Their level of significance (p) for 2 x 2 comparisons was determined as previously described [19, 20]. In addition, the most frequent complete extended haplotypes were deduced from: 1) the 2, 3, and 4 HLA loci haplotype frequencies [19, 20]; 2) the previously described haplotypes in other populations [19, 20]; and 3) haplotypes if they appeared in two or more individuals and the alternative haplotype was well defined [19, 20]. In order to compare phenotype and haplotype HLA frequencies with other populations, the reference tables of the 11th and 12th International HLA Workshops were used [21, 22]. Phylogenetic trees (dendrograms) were constructed with the allelic frequencies using the Neighbour-Joining (NJ) method [23] with the genetic distances between populations (DA) [24], using DISPAN software comprising the programs GNKDST and TREEVIEW [25, 26]. Correspondence analysis in three dimensions and its bidimensional representation was carried out using the VISTA v5.05 computer program [27]. Correspondence analysis consists of a geometric technique that may be used for displaying a global view of the relationships among populations according to HLA (or other) allele frequencies. This methodology is based on the genetic distances (DA) variance among populations (similar to the classical principal components methodology) and of a statistical visualization of the differences.
RESULTS
HLA Allele Frequencies Found in Kurd Population: Comparisons with Other Populations
The expected and observed gene frequency values for HLA-A, -B, -DRB1, and -DQB1 loci do not differ significantly and the population is found in Hardy-Weinberg equilibrium (data not shown). Table 2 shows the HLA allele frequencies found in the sampled population. Twenty-one different HLA-A and twenty-nine different HLA-B alleles were found in Kurd sample (Table 2). Only nine HLA-A alleles and eleven HLA-B alleles had frequencies higher than 4% (A*01:01, A*02:01, A*02:50, A*02:52, A*03:01, A*11:01, A*24:02, A*26:01, A*32:01, B*08:01, B*18:01, B*35:01, B*38:01, B*44:02, B*49:01, B*50:01, B*51:01, B*52:01, B*55:01, and B*58:01). Seventeen different HLA-DRB1 and fourteen different HLA-DQB1 alleles were found (Table 2). Ten HLA-DRB1 and nine HLA-DQB1 alleles had frequencies higher than 4% (DRB1*01:01, DRB1*03:01, DRB1*04:01, DRB1*07:01, DRB1*11:01, DRB1*13:01, DRB1*13:02, DRB1*14:01, DRB1*15:01, DRB1*16:01, DQB1*02:01, DQB1*03:01, DQB1*03:02, DQB1*03:05, DQB1*05:01, DQB1*05:02, DQB1*06:01, DQB1*06:02 and DQB1*06:03).
Two types of analyses were done in order to compare Kurd HLA frequencies with other World population frequencies: 1) by using pooled DRB1 and DQB1 data, and 2) by using DRB1 only. It was not possible to carry out a study comparing HLA class I allele frequencies or HLA class I and II conjointly due to the lack of class I studies in many worldwide populations, see Table 1. Thus, a single DRB1 study was also carried out in order to compare the Kurd HLA population frequencies with those of as many as possible populations. The NJ relatedness dendrogram based on HLA-DRB1 allele frequencies (Fig. 2) separates populations in two well-differentiated clusters. One of them groups North and South Mediterraneans (Europeans and Africans), Middle Easterns (included Kurds) and western Siberians. The second cluster grouped the rest of analyzed populations (central and eastern Siberians and Orientals). Kurds are integrated in the first cluster, together with Gorgan (Iranian Turkmen population [28]), Iranian populations (specially from Fars province [1]), Russian Chuvash (who live near lower Volga River, North Caspian Sea [29]), Russian Siberian Mansi (from western Siberia [30]), and Russian-Mongols Buryat (from Baikal Lake region [30]). Correspondence analyses based on HLA-DRB1 allele frequencies (Fig. 3) and on HLA-DRB1 and HLA-DQB1 frequencies conjointly (data not shown) show identical results. Also, two groups are clearly defined according to first dimension that explains most of the variability among populations. The first one includes Oriental and Siberian populations (together with Gorgan Turkmen) and the second one is divided according to the second dimension into two subgroups. First subgroup clusters Europeans, northern Mediterranean, Caucasus and Iranian populations, while second subgroup clusters southern and eastern Mediterraneans; Kurds are located in an intermediate situation between these two major groups in this analysis but closer to the second one (Fig. 3).
This effect is more evident in Table 3, which show that Kurds closest genetic distances are the following: Gorgan population, Caucasoid European Mediterranean groups such as Russian, Italians, Croatians, Germans and Turks, populations from Caucasus such as Georgian and Svan, Iranians from Fars province, and Chuvash from western Siberia.
HLA-A, -B, -DRB1 and -DQB1 Extended Haplotype Analysis in Kurds: Comparison with Other Populations
Associations between different HLA loci were estimated in Kurds. The most probable two loci haplotypes (A-B, B-DRB1 and DRB1-DQB1) were calculated. These two-loci linkage disequilibrium data (not shown) show that the most frequent combinations are characteristic of Siberian or Central Asian populations and they have found in several Siberian populations (DRB1*11:01-DQB1*03:01, DRB1* 01:01-DQB1*05:01, DRB1*07:01-DQB1*02:01, DRB1* 15:01-DQB1*06:01) [3, 29-32]. However, several two-loci combinations found in this population in a considerable high frequency are characteristic of Middle East and Mediterranean populations (A*02:01-B*50:01, DRB1* 03:01-DQB1*02:01) [1, 33]. The twelve most frequent four HLA loci haplotype combinations (A-B-DRB1-DQB1) were calculated and these extended haplotypes are depicted in Table 4; they represent 26.8 % of all haplotypes. Class I haplotype A*02:01-B*35:01 is present in association with DRB1*01:01-DQB1*05:01 and DRB1*11:01-DQB1*03:01. This class II haplotype DRB1*11:01-DQB1*03:01 is also associated with A*24:02-B*18:09, A*01:01-B*35:01, and A*03:01-B*44:02, and other class II haplotype, DRB1*03: 01-DQB1*02:01, is present in association with A*24:02-B*58:01, A*01:01-B*08:01, and A*02:01-B*35:01. The Kurd extended HLA haplotypes obtained allow their comparison with previously reported ones in other populations (Table 4 and its footnote). In summary, most of two-loci haplotypes and extended haplotypes are either from Asian-Siberian or Middle East-Mediterranean origin. New found extended HLA haplotypes in Kurds are also quoted (Table 4).
DISCUSSION
Genetic distances studies (both NJ relatedness tree and correspondence analysis) place Kurds in the Mediterranean cluster, close to Russians, Southern Russian Chuvash and Caucasus populations (Svan and Georgians) (Figs. 2, 3). Furthermore, correspondence analysis shows Kurds close to Iranian populations like Baloch and Iranians from Fars, and Gorgan Turkmen (the closest population according to plain genetic distances), in a half-way position between Mediterraneans and Western and Central Siberians, together with Chuvash. In fact, Asian and Mediterranean traits were found Russian Chuvash HLA profiles in a previous study [29]. One of the most frequent Kurd HLA extended haplotype found in this study, A*01:01-B*08:01-DRB1*03:01-DQB1*02:01, is shared with Chuvash.
New extended HLA haplotypes are found in Kurds: A*02:01-B*35:01-DRB1*01:01-DQB1*05:01, A*24:02-B* 18:09-DRB1*11:01-DQB1*03:01, A*03:01-B*44:02-DRB1 *11:01-DQB1*03:01, A*24:02-B*52:01-DRB1*15:01-DQB 1*06:01, A*24:02-B*58:01-DRB1*03:01-DQB1*02:01, A* 02:01-B*35:01-DRB1*11:01-DQB1*03:01, A*02:01-B*50: 01-DRB1*07:01-DQB1*02:01, A*11:01-B*50:01-DRB1* 07:01-DQB1*02:01, A*24:02-B*51:01-DRB1*08:01-DQB1 *04:01 and A*26:01-B*08:01-DRB1*03:01-DQB1*02:01. Some of the HLA class I or class II splits of these extended haplotypes are shared with other populations (mainly Mediterraneans and Siberians), but none of them with Iranian populations from Yazd and Shiraz. This fact suggests that differences exist may with old Iranian surrounding populations and make this study worthwhile for establishing future regional transplant programs for Kurds. Also HLA and disease epidemiology studies [34] are addressed with the present study results, together with specific HLA pharmaco-genomics for this particular population. Different therapeutic drugs may differentially affect Kurds according to this paper defined HLA profile [4].
Kurds and Turkish speak languages belonging to different families [35]. However, HLA genetics distances observed in Kurds put them into Mediterranean stock together with Turkish (Table 3, Figs. (2, 3). Genetic studies based on Y-chr in Kurd groups from several regions (Turkey, Georgia and Iran) identify the dominant presence of haplogroups originated in Middle-East (Anatolia or Mesopotamia) showing a close association with Jews, Lebanese and Turkish genes and languages do not correlate [7, 8, 36]. The origin of Iranian peoples as a hypothetical Aryan (Indo-Iranian) invasion has been very much debated. The invasion by these tribes is only based on linguistics, so the fact that this invasion existed as an “indoiranization” process through an “elite” of rulers that imposed their culture, included language, is strongly possible. Soon after of the beginning of the 1st millennium B.C. (1000-900 B.C.) Medes came to notice as an empire conquering the Zagros mountains and lower Mesopotamia [37]. The Persian Achaemenid dynasty that conquered the Median Empire, also with an Indo-Iranian origin, expanded westwards and took over most of Middle East, including Anatolia and Egypt. Macedonian Alexander the Great brought it to an end in the 4th century B.C establishing the Seleucid dynasty. Later, Arabs also brought to an end the Sasanian (Persian) Empire by 638 A.D. Old Persian language survived [38]. Thus, it is likely that nowadays Iranians were settled down in their area since prehistorical times.
In spite of the fact that Kurds do not share any of the most frequent extended haplotypes with Iranians, plain chord genetic distances between them are close (Table 3). Previous HLA studies show that modern Iranians are close to other Middle East-Mediterranean populations (Macedonians, Cretans and Turkish) [6, 39]. Thus, this HLA genetic data support the hypothesis that present day Iranian genetic stock comes from the ancient autochthonous people and not from hypothetical “Aryan” invasion from East [5, 6].
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
ACKNOWLEDGEMENTS
This work was supported in part by grants from the Spanish Ministry of Health (FISS PI080838 and PI11/00807) and three different Mutua Madrileña Automovilista grants.


