All published articles of this journal are available on ScienceDirect.
HLA-G in Amerindians: Epidemiology and Worldwide Population Comparison
Abstract
Background:
HLA-G molecules are immunosuppressive and avoid fetal rejection by giving negative signals to maternal immune system from fetal trophoblast cell surface. HLA-G genes have been associated to different pathologies: Spontaneous abortions, autoimmunity, tumor progression, transplant rejection and infection. In addition, different World populations show remarkable different HLA-G allele frequencies in the allele that does not produce a full HLA-G molecule (HLA-G*05N); this allele is almost absent in studied Amerindians.
Objectives:
The aim is to study HLA-A.-B,-DRB1 and –G alleles and extended haplotypes in Amerindians for the first time. This may be useful to asses HLA-G epidemiology, association to disease and Preventive Medicine in Amerindians.
Methods:
HLA-A,-B and -DRB1 have been typed by using standard automatic protocols. HLA-G alleles have been detected by direct HLA-G exon 2, exon 3 and exon 4 DNA sequencing. Computer calculations have been done by specific standard methods.
Results:
HLA-A,-B,-DRB1 and –G extended haplotypes have been calculated in Amerindians for the first time. Also, their HLA-G frequencies have been compared with worldwide populations.
Conclusion:
Low frequencies of null HLA-G*01:05N allele are found in Amerindians. The extended haplotypes with this allele bear other typical Amerindian HLA-DRB1 alleles and its origin is discussed. HLA-G allele frequency profile is closer to that of Europeans than to that of Far East Asians. Our findings are useful to Preventive Medicine and Epidemiology associated to Fertility and HLA-G associated pathology and transplantation.
1. INTRODUCTION
The First Native Americans are postulated to have populated America coming from Asia through the Bering land bridge between 30,000 and 12,000 years before present (BP). Siberia [1] and Mongolia [2, 3] have been thought as the most approximate places of Amerindians origin in Asia. A study [4] proposed the triple migration hypothesis for explaining the Americas peopling on linguistic bases: Amerindians (most North and South American Indians; 12,000 BP), Na-Dene (Athabascans, Navajo, Apache; 8000 BP) and Eskimo-Aleuts (6000 BP). However, Y chromosome markers established that more than one paternal founder haplotype arrived into America on different migration times [5, 6]. Alu insertions (also nuclear DNA) research [7]: suggest that the geographical distribution was the result of only one wave of immigration. In addition, East Asian HLA genes have been found in the Azores Islands [8]; is an evidence of spreading of Chinese fleet around the world in 1421 [9]. Also, South American peopling may have come from Asia or Polynesia because HTLV-1 virus strain shared identical DNA sequences in Japan and in the northern coast of South America [10] and some HLA alleles may have been exchanged by a Trans-Pacific route [11-14]. Both genetic [8] and archaeological [15] evidence suggest that two-way Trans-Atlantic traffic occurred before Europeans came to America in 1492 AD. Tools in New Mexico have recently found used 20,000 years ago in Spanish Solutrean culture [13-16]. Other prehistoric common artifacts to America and Europe have been described [17].
On the other hand, HLA-G proteins have been found to be tolerogenic MHC molecules. They were described being expressed in placental cytotrophoblast and have a role in fetal/maternal immunological tolerance [18] and also in disease like spontaneous abortions, tumor immunology, transplantation and autoimmunity [19]. However, HLA-G is also expressed normally in other tissues comprising cornea, adult thymic epithelial cells, erythroblasts, pancreatic islets, endothelial precursors and mesenchymal stem cells [20]. Its polymorphism is low compared to classical class I loci [21]. HLA-G surface protein has interactions with cell receptors of the immune system, including the killer cell immunoglobulin-like receptor KIR2DL4, CD8, LILRB1 and LILRB2 [22]. HLA-G*01:05N (first described in Spaniards [23]) is being maintained throughout time in World populations [24] and it was shown that homozygous HLA-G*01:05N females can normally give birth and that HLA-G*01:05N individuals are born without problems, and do not show at 30–40 years old any symptom of immunodeficiency, autoimmunity or tumor pathology [25].
Most Amerindian XXI century immigrants in Madrid come from Andean Countries (mainly Ecuador, Colombia, Peru and Bolivia). They show an HLA gene profile with “quasi-specific alleles”, which makes them different to the rest of the World, and they never had been typed for HLA-G gene before. Recently, immigrated volunteer unrelated blood donors contributed to the present study [26].
Our aims are: 1) To analyze HLA-G allele frequencies in Amerindian immigrant healthy population to be used as a documented reference for future studies on HLA-G population variation allele frequency and its association with unexplained infertility and other disorders; 2) To analyze four loci (HLA-A, -B, -DRB1, -G) extended haplotypes frequencies and compare them with those of other worldwide populations in order to study the HLA relatedness with peoples most likely to be candidates for First American Peoples ancestors; 3) To analyze HLA-G*01:05N allele frequencies in our population in order to a precise view of HLA-G*01:05N allele frequency distribution throughout different World populations. Our results may be important in Amerindian-infections Epidemiology studies, their Preventive Medicine of fetal/mother normal and pathological relationships and other autoimmune, tumor or transplant pathologies.
2. MATERIALS AND METHODS
2.1. Population Sample
154 healthy unrelated Amerindian and Caribbean individuals were HLA-G typed [26]. Samples were collected from volunteer immigrant Amerindian blood donors at The Madrid Regional Blood Center. A written consent to participate in the present study was signed by each blood donor. Individuals were classified as Amerindian both by physical anthropology and by born place in an Andean or other Iberian-American country and their Amerindian origin was also assessed confirmed by full typing of HLA-A, -B, DRB1 and –DQB1 [11]. Only individuals who come from rural or tribal areas and whose grand-parents come from the same areas were used for the study. Individual country origin and number were the following ones: Ecuador, 45; Colombia, 42; Peru, 21; Bolivia, 10; Argentina, 7; Dominican Republic, 6; Brazil, 5; Mexico, 4; Uruguay, 4; Chile, 4; Paraguay, 2; Venezuela, 2; Honduras, 1; Nicaragua, 1; Cuba, 1 and Guatemala, 1.
2.2. DNA Extraction, Amplification and HLA-G Sequencing
Genomic DNA from peripheral blood nucleated cells was extracted with the Nucleic Acid Isolation System QuickGene-810 (Fujifilm, Japan) by using reagents and protocols provided by the manufacturer (QuickGene Whole Blood Extraction Kit S, Fujifilm, Japan).HLA-A,-B,-DRB1 alleles typing were obtained by a standard protocol [14]. HLA DNA amplification of exons 2, 3 and 4 of HLA-G was carried out separately by direct PCRs. In order to obtain each HLA-G exons sequences, the following specific 5’ and 3’ primers were used. Exon 2: forward: 5’-GAGGGTCGGGCGGGTCTCAAC-3’, reverse: 5’-GCATGGAGGTGGGGGTCGTGA-3’; Exon 3: forward 5’-TGGGCGGGGCTGACCGAGAAGGTGG-3’, reverse: 5’-CTCTCCTTGTGCTAGGCCAGGCTGAGA-3’; Exon 4: forward: 5’-CCATGAGAGATGCAAAGTGCT-3’, reverse: 5’-TGCTTTCCCTAACAGACATGA-3’. The PCR conditions consisted of 1 cycle of 95°C, 15 min; 95°C, 0.5 min; 35 cycles of 68°C, 0.5 min; 72°C, 0.67 min and 1 cycle of 72°C, 10 min for the exon 2; 1 cycle of 94°C, 2 min; 94°C, 1 min; 35 cycles of 67°C, 1.5 min; 72°C, 2 min and 1 cycle of 72°C, 10 min for the exon 3; and 1 cycle of 95°C, 5 min; 95°C, 0.67 min; 35 cycles of 58°C, 0.67 min; 72°C, 0.67 min and 1 cycle of 72°C, 10 min for the exon 4, in a programmable heat block (Mastercycler epgradient S, Eppendorf, Germany). The obtained products were electrophoresed in a 2% agarose and detected by staining with Olerup SSP Gelred and were purified by using the Mini Elute Gel Extraction Kit 250, QIAGEN. Purified products were DNA sequenced using the ABI 3730 DNA analyzer (Applied Biosystems, Foster City, CA, USA) by using the same primers of amplification process. HLA-A,-B,-DRB1 typings were obtained by a standard protocol [14]. DNA HLA-G polymorphism was identified by aligning the sequences with identified alleles [21] using Mega 5 software [27].
3. RESULTS
3.1. HLA-G allele Frequencies in Amerindian Population from Madrid
18 of the 51 HLA-G alleles described [21] are present in our Amerindian population (Table 1). The three most frequent alleles are: HLA-G*01:01:01 (31.2%); HLA-G*01:01:02 (23.1%) and HLA-G*01:04:01 (12.8). The HLA-G*01:05N null allele is present in a frequency of 1.3%.
| HLA-G alleles | Allele Frequency (%) | HLA-G alleles | Allele Frequency (%) |
|---|---|---|---|
| HLA-G*01:01:01 | 31.2 | HLA-G*01:01:19 | 0.4 |
| HLA-G*01:01:02 | 23.1 | HLA-G*01:01:20 | 1.3 |
| HLA-G*01:01:03 | 4.3 | HLA-G*01:03:01 | 7.7 |
| HLA-G*01:01:05 | 0.9 | HLA-G*01:04:01 | 12.8 |
| HLA-G*01:01:06 | 2.1 | HLA-G*01:04:03 | 1.7 |
| HLA-G*01:01:07 | 0.4 | HLA-G*01:04:04 | 3.8 |
| HLA-G*01:01:08 | 3.0 | HLA-G*01:04:05 | 0.4 |
| HLA-G*01:01:15 | 0.4 | HLA-G*01:05N | 1.3 |
| HLA-G*01:01:17 | 0.9 | HLA-G*01:06 | 0.4 |
3.2. Haplotypes in Amerindian Population from Madrid
Frequencies of two loci (HLA-A-B) and four loci (HLA-A-B-DRB1-G) HLA haplotypes were calculated. Results are shown in Tables 2 and 3. Results of the closest HLA studied locus to HLA-G (i.e.: HLA-A) are shown in Table 2.
| HLA-A-G | HF (%) | HLA-A-G | HF (%) |
|---|---|---|---|
| A*02:01-G*01:01 | 24.6 | A*01:01-G*01:06 | 3.3 |
| A*24:02-G*01:04 | 13.2 | A*68:02-G*01:01 | 2.9 |
| A*03:01-G*01:01 | 6.6 | A*32:01-G*01:01 | 2.9 |
| A*68:01-G*01:01 | 5.2 | A*30:02-G*01:01 | 2.4 |
| A*29:02-G*01:01 | 5.0 | A*26:01-G*01:01 | 2.4 |
| A*23:01-G*01:04 | 3.3 | A*30:01-G*01:05N | 0.8 |
| A*11:01-G*01:01 | 3.3 | A*31:01-G*01:05N | 0.4 |
| A*02:11-G*01:01 | 3.3 |
The 13 two loci most frequent HLA-A-G haplotypes are depicted in Tabel 2. Two of the haplotypes contain the null allele HLA-G*01:05N. A*30:01-G*01:05N goes either with B*45:05 or B*45:01 and A*31:01-G*01:05N goes with B*15:05. A*02:01-G*01:01 (24.6%) and A*24:02-G*01:04 (13.2%) are the two loci haplotypes with higher frequencies.
| HLA-A-B-DRB1-G | HF (%) | Also found in |
|---|---|---|
| A*24:02-B*35:01-DRB1*14:02-G*01:04a | 2.9 | Amerindian |
| A*02:01-B*35:04-DRB1*08:02-G*01:01b | 1.7 | Amerindian |
| A*02:01-B*40:02-DRB1*04:07-G*01:01c | 1.2 | Amerindian |
| A*03:01-B*07:02-DRB1*15:01-G*01:01d | 1.2 | North Africans,Europeans |
| A*02:01-B*44:03-DRB1*13:02-G*01:01e | 1.2 | Italian, German, Japanese |
| A*03:01-B*51:01-DRB1*04:04-G*01:01f | 1.2 | German |
| A*24:02-B*14:02-DRB1*07:01-G*01:01g | 1.2 | German |
| A*68:01-B*40:04-DRB1*04:03-G*01:01h | 1.2 | Venezuelan (probably Amerindian) |
| A*31:01-B*39:06-DRB1*13:02-G*01:01i | 1.2 | New |
| A*29:02-B*15:05-DRB1*04:03-G*01:05Nj | 0.6 | New |
| A*31:01-B*15:05-DRB1*04:03-G*01:05Nk | 0.6 | New |
| A*33:03-B*57:03-DRB1*13:02-G*01:05Nl | 0.6 | New |
The most frequent four loci haplotype in our Amerindian population is A*24:02-B*35:01-DRB1*14:02-G*01:04 with a frequency of 2.9% (Table 3). This haplotype is found without HLA-G locus in other populations, such as, Aymara, Quechua and Jaidukama (see Table 3 footnote for references). Two alleles of this haplotype, HLA-A*24:02 and HLA-G*01:04 are in linkage disequilibrium in our studied population ( Table 2). The second most frequent four loci haplotype is A*02:01-B*35:04-DRB1*08:02-G*01:01 with a frequency of 1.7% (Table 3). This haplotype without HLA-G locus is also found in other Amerindian populations, such as, Mayan, Aymara, Quechua, Mayos, Nahua and Wayu (see Table 3 footnote for references). Two alleles of this haplotype, HLA-A*02:01 and HLA-G*01:01, are in linkage disequilibrium (data not shown) in our population.
In Table 3, it can be observed that the most frequent haplotypes without HLA-G locus have a possible Amerindian origin. There is a certain European influence, that can be observed in the haplotype A*03:01-B*07:02-DRB1*15:01-G*01:01, since this haplotype has a possible European or North African origin and has a high frequency in North Spain populations [40] and Irish populations [41]. The first most frequent Amerindian four loci extended haplotypes bear HLA-G*01:04 allele. The last four haplotypes A*31:01-B*39:06-DRB1*13:02-G*01:01, A*29:02-B*15:05-DRB1*04:03-G*01:05N, A*31:01-B*15:05-DRB1*04:03-G*01:05N and A*33:03-B*57:03-DRB1*13:02-G*01:05N have not been found in any other populations (see Table 3 footnote for details).
3.3. Null Allele, HLA-G*01:05N, in Amerindian Population from Madrid
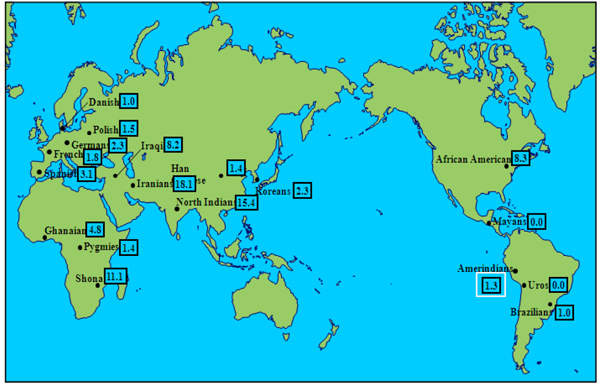
Three null alleles were found in three different individuals, each of them from a different country, i.e.: Colombia, Argentina and Nicaragua. The HLA-G*01:05N frequency was 1.3% as it shows in Table 1. Other populations that show low frequency of null allele HLA-G*01:05N are mixed Brazilians, Amerindian Uros and Amerindian Mayans. On the other hand, the highest null allele frequencies are observed in Middle East populations, such as, North Indians, Iranians and Iraqi and in African populations except Pygmies, such as, Ghanaian, African American and Shona. These six populations that present high frequencies of null allele, have significant frequency differences compared with our studied population of Amerindians from Madrid by Fisher Test with a p value <0.05.
4. DISCUSSION
4.1. HLA-G Polymorphism in Our Amerindian Sample
A characteristic of HLA-G is its low polymorphism. It is suggested that it could be due to its tolerogenic and not presentation function. Most of HLA-G alleles have been found to be silent mutations in exons/introns [41, 42]. However, many of HLA-G alleles have been described only in one study, since they have not been found in other populations [41]. Asian, African and Caucasoid populations have a low HLA-G allelic variability compared with the rest of classic class I HLA genes [41, 43]. Amerindian populations have in common the limited number of classic class I HLA alleles [38]. Regarding to non classic class I HLA alleles, as HLA-G, there are only a few studies about their frequencies in these populations [41, 44, 45].
Only four HLA-G different proteins are usually typed in World populations. These proteins are encoded by most frequent groups of HLA-G alleles, such as, HLA-G*01:01, G*01:03, G*01:04 y G*01:06, which could be different regarding HLA-G function [19]. Eighteen different DNA alleles have been detected in the studied population, being the most frequent HLA-G*01:01:01 with a frequency of 31.2%. This low numbers are concordant to those found in Brazil, China, Ghana, Poland and Korea and others [41]. Eleven different alleles within the HLA-G*01:01 group of alleles, have been, while four alleles have been detected in HLA-G*01:04 group (see Table 1). In other populations the number of HLA-G alleles is very limited (Koreans [46], Shona African [47] and Danish [48]). However, the number of alleles in our population is relatively high as also found in Brazilians [45]. The lack of worldwide exchange and international HLA-G workshops may be the cause of the observed low polymorphism and in the future HLA-G may show to be as polymorphic as other HLA class I locus.
Two new HLA-G alleles have been described in our studied Amerindians sample [49]. One new allele was found in a male individual from Ecuador, with the following GenBank accession number: KP739973; it was named HLA-G*01:08:02 by the IMGT/HLA database [21, 49, 50]. The other new allele was found in a female individual from Hispaniola Island (Sto Domingo); its GenBank accession number is KP739974. The IMGT/HLA database named it as HLA-G*01:20 [21, 49, 50].
4.2. Analyses of Extended Haplotypes in Amerindian Population
In the present study, four loci (HLA-A-B-DRB1-G) extended haplotype have been calculated (see Table 3). The most frequent extended haplotypes in the studied population are shown in Table 3. The three first are A*24:02-B*35:01-DRB1*14:02-G*01:04, A*02:01-B*35:04-DRB1*08:02-G*01:01 and A*02:01-B*40:02-DRB1*04:07-G*01:01 with a frequency of 2.9%, 1.7% and 1.2% respectively. These haplotypes without HLA-G locus are characteristic of Amerindian and have also been observed in Aymara [31], Quechua [32], Jaidukama [33], Mayan [35], Nahua [37], Wayu [36], Lama [38] and Mayo [34] populations. Two of these haplotypes, A*24:02-B*35:01-DRB1*14:02-G*01:04 and A*02:01-B*35:04-DRB1*08:02-G*01:01 bear DRB1*08:02 and DRB1*14:02 alleles which are considered “quasi specific” markers of Amerindian populations [51, 52]. In addition, haplotype A*02:01-B*40:02-DRB1*04:07-G*01:01 bears DRB1*04:07 allele, which is also observed in Amerindians [38].
The Amerindian HLA profile is different compared to the rest of World [11-14, 53-55], and it is confirmed in our present study; HLA-A*03:01-B*07:02-DRB1*15:01-G*01:01 haplotype has a possible North African or European origin (see Table 3). This haplotype has been found without HLA-G in high frequency in North Spain populations [40] and also in northern European populations [41].
The last four extended haplotypes in Table 3 have been not been found in any other population.
4.3. HLA-G*01:05N in Amerindians: Comparison with Worldwide Populations
Three null alleles were detected in three different individuals, all of them heterozygous (Tables 1 and 2). In 1997, the first null allele of HLA-G locus was described by us in a Spanish population sample [23]. This null allele permitted transcription and translation of HLA-G isoforms bearing only alpha-1 domain. Exon 3 is removed by a previous DNA base deletion and a consequent shifting of reading frame which generates a stop codon [19, 23, 56]. Frequencies of Uros (from the Titikaka Lake) and Mayas (from Guatemala) null allele were very similar to those found now by us. This null allele has been associated with spontaneous and recurrent abortions and, in the other hand, also with a protective effect against intrauterine infections during the gestation [19, 44]. The absolute absence of null allele both in Uros and Mayas contradicts the hypothesis that frequent intrauterine infections can maintain null allele frequencies, since health care in Amerindian communities is and has been poor, comparatively to European ones. Also, Amerindian admixed Brazilian population show similar results [45]. Populations with a significantly higher HLA-G*01:05N frequency include some Middle East Caucasians (Iraqis, Iranians, Indians from North India) and certain African populations (Ghana, Shona and African Americans) (Fig. 1).
HLA-G*01:05N allele DNA sequence may indicate that it was originated from the HLA-G*01:01:02 allele, following the deletion of the cysteine at the 129/130 codon, since its sequences are identical, except for such deletion [25]. In addition, HLA-G* 01:05N is associated with the HLA-A*30:01-B*13:02 haplotype prevalent in the Middle East and some Mediterranean populations, as it may have been introduced in Spain by Berber/Muslim invaders in the 8th century AD [13, 14, 23], or when the Mediterranean glacial refugee was established around 10,000 BC [65, 66], which could place the Middle East as the origin of this mutation [44].
Regarding the extended haplotypes studied, three of the entire list of haplotypes obtained carried the null allele; HLA-A*33:03-B*57:03-DRB1*13:02-G*01:05N, HLA-A*31:01-B*15:05-DRB1*04:03-G*01:05N and HLA-A*29:02-B*15:05-DRB1*04:03-G*01:05N (Table 3), which still shows the low presence of this allele in Amerindian populations. These three haplotypes have not been found in any other population (Table 3 and footnote). They are in part Amerindian since DRB1 alleles were typical Amerindian [11]. HLA-B*15:05 allele has a possible Amerindian origin [41], and two of the extended haplotypes carry this allele, HLA-A*31:01-B*15:05-DRB1*04:03-G*01:05N and HLA-A*29:02-B*15:05-DRB1*04:03-G*01:05N (Table 3). HLA-B*57:03 allele has a possible African, Caucasoid origin [41] and is it observed in the extended haplotype HLA-A*33:03-B*57:03-DRB1*13:02-G*01:05N. Linkage disequilibrium may be mainly indicative of a population admixture in that region [67], so it could be understood that there has been a low degree of admixture in the studied Amerindian population, probably with Europeans and Africans.
The existence of “diseased” genes in a population is often justified by evolutionary advantages. As discussed above, the HLA-G immunosuppressive molecule plays a major role in the fetus’s acceptance of the mother during gestation [19]. The maternal-fetal interests may become contradictory due to lower levels of HLA-G, the mother's immune system could cause fetal damage and finally spontaneous abortions in heterozygous or healthy homozygous mothers for the HLA-G*01:05N allele [44]. However, it was observed that women and fetuses of the same Spanish family, homozygous for the HLA-G*01:05N allele, were healthy individuals capable of having completely normal deliveries and being born, indicating that the HLA-G1 isoform is not essential for survival [25]. This is further supported by the findings that genus Macacca primates survive healthy with HLA-G incomplete molecules, due to a DNA deletion [68, 69]. Protective effects of this allele have been described against intrauterine infections in pregnant women. It is unclear, but both circumstances could coexist [44]. The presence of this null allele in the population could be due to the fact that the α1 domain is sufficient for the normal functioning of the HLA-G molecule, so negative evolutionary forces would not act to eliminate this gene. Other advantages than avoiding maternal rejection of fetus may be attributed to this null allele: HLA-G*01:05N allele also may provide some selective advantage, not only maternal-fetal, but to improve the level of immune response against HIV infection [70] or other infections of the immune system not directly related to pregnancy.
4.4. HLA-G*01:04 and HLA-G*01:01 Allele Frequencies in Amerindians and Worldwide Populations
The highest frequencies of HLA-G*01:04 allele are found in Japanese, Iranian and South Korean populations (45%, 29.4% and 27.7% respectively) (Fig. 2). Our Amerindian population has a frequency of 18.7%. Other Amerindian populations have similar frequencies to that found in our present sample, such as Uros from Titikaka Lake with a frequency of 10.2% and Mayans from Guatemala with a 13.1%. It is important to highlight that has not been detected any population with a frequency of this allele higher than 10% in central Europe and higher than 13% in Iberia (Portuguese 13%, Spaniards 11%, (Fig. 2). Significant HLA-G frequency differences have not been found between Amerindians and the rest of World populations (results not shown), but only a trend to lower frequency in central Europe is observed.
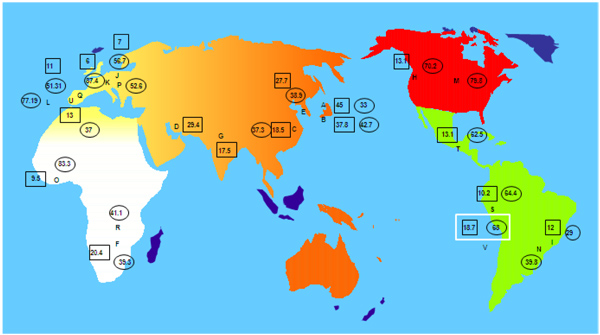
A- Japan pop 14: G*01:04→ 45%, G*01:01→ 33% [71]
B- Japan pop 15: G*01:04→ 37.8%, G*01:01→ 42.7% [64]
C- China Zhejiang Province Han: G*01:04→18.5%, G*01:01→ 37.3% [63]
D- Iran Tehran pop 2: G*01:04→ 29.4% [58]
E- South Korea pop 7: G*01:04→ 27.7%, G*01:01→38.9% [72]
F- Zimbabwe Harare Shona pop 2: G*01:04→20.4%, G*01:01→ 39.3% [73]
G- India Lucknow pop 2: G*01:04→ 17.5% [57]
H- USA African American pop 6: G*01:04→ 13.1%, G*01:01→ 70.2% [64]
I- Brazil Parana mixed: G*01:04→ 12%, G*01:01→ 29% [74]
J- Denmark pop2: G*01:04→ 7% [75], G*01:01→ 56.7% [76]
K- Germany pop5: G*01:04→ 6%, G*01:01→ 87.4% [60]
L- Portugal Azores Terceira Island: G*01:01→ 77.19% [41]
M- USA South Dakota Hutteritie: G*01:01→ 79.8% [77]
N- Brazil Sao Paulo pop 2: G*01:01→ 39.8% [45]
O- Ghana: G*01:04→ 9.5%, G*01:01→ 83.3% [64]
P- Poland pop 2: G*01:01→ 52.6% [62]
Q- Spain pop 2: G*01:04→ 11%, G*01:01→ 51.31% [23]
R- Zambia: G*01:01→ 41.1% [78]
S- Titikaka Lake Uros: G*01:04 → 10.2%, G*01:01→ 64.4% [44]
T- Guatemala Mayans: G*01:04→ 13.1%, G*01:01→ 62.5% [44]
U- Portuguese: G*01:04→13%, G*01:01→ 37% [79]
V- Amerindians from Madrid: G*01:04→ 18.7%, G*01:01→ 68% (present study)
Regarding to [2] HLA-G*01:01 allele, the highest frequencies are found in Germany, Ghana and USA South Dakota Hutteritie populations (87.4%, 83.3% and 79.8% respectively). Our studied Amerindian population has a frequency of 68%, very similar to other Amerindian populations, such as, Uros from Titikaka Lake and Mayans from Guatemala, (64.4% and 62.5% respectively). Significant differences have not been found between Amerindians and the rest of World populations (results not shown), only a trend to a higher frequencies in Germany and Africa (Ghana) (Fig. 2).
CONCLUSION
Present study results may be important in the field of epidemiology studies of infection, abortions, infertility and fetal pathology and Preventive Medicine at the maternal-fetal relationship.
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
Not applicable.
HUMAN AND ANIMAL RIGHTS
No Animals/Humans were used for studies that are base of this research.
CONSENT FOR PUBLICATION
Not applicable.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
This work was supported in part by Grants from the Spanish Ministry of Health and Economy PI14/01067 and European FEDER funds. We declare that these results have been fully obtained in our department without any external conflict of interest.


