All published articles of this journal are available on ScienceDirect.
TRIM65 Promotes Osteogenic Differentiation by Regulating the PI3K/AKT Signaling Pathway
Abstract
Introduction
Tripartite motif 65 (TRIM65) is a crucial regulator of cell differentiation, proliferation, migration, invasion, and carcinogenesis. However, its role in osteoporosis (OP) remains unclear. In this study, we evaluated the role of TRIM65 in regulating osteoblast differentiation and calcification.
Materials and Methods
The role of TRIM65 during the osteogenic differentiation of MC3T3-E1 cells was evaluated. The expression of COL1A1, RUNX2, and OCN was examined using western blot analysis and immunofluorescence staining. The formation of calcium nodules was evaluated using alizarin red staining. Alkaline phosphatase activity was evaluated using ALP staining.
Results
TRIM65 expression was significantly elevated during the osteogenic differentiation of bone marrow mesenchymal stem and MC3T3-E1 cells. We demonstrated that TRIM65 overexpression enhanced osteogenic differentiation and promoted bone formation in the MC3T3-E1 cells. Conversely, TRIM65 inhibited the osteogenic differentiation and bone formation of the MC3T3-E1 cells. Mechanistically, we found that TRIM65 knockdown in MC3T3-E1 cells up-regulated the phosphorylated protein expression of PI3K and AKT, which was contrary to the results of the TRIM65-overexpression group.
Conclusion
Our research suggests that TRIM65 is an important osteogenic differentiation and bone formation regulator and offers a therapeutic application for OP.
1. INTRODUCTION
Osteoporosis (OP) is a systemic bone disease characterized by the destruction of bone microstructure, decreased bone mass, and increased susceptibility to fracture [1-5]. The prevalence of osteoporotic fractures globally is 8.9 million annually [6-10]. More than 200 million people have been globally diagnosed with OP [11-13]. Osteoporotic fractures are a growing public health concern, especially among postmenopausal elderly women [14-17]. Therefore, finding an effective treatment strategy to diagnose and treat osteoporosis is important. In 1994, the WHO recommended that bone mineral content, which is maintained by a dynamic balance between bone resorption and formation, is the main standard for OP diagnosis [18]. Osteoblasts are the major functional cells of bone formation, responsible for the synthesis, secretion, and mineralization of bone matrix and the stages of differentiation. Proliferation and mineralization are controlled by diverse transcription factors, cytokines, and signaling pathways [19-21]. Impaired osteoblast differentiation, proliferation, and mineralization are the major pathophysiological mecha- nisms underlying OP [22, 23]. However, the underlying mechanisms remain obscure. Therefore, further elucidation of the regulatory mechanisms of osteoblast differentiation and mineralization is required for under- standing OP pathogenesis and offers new strategies for its treatment.
Tripartite motif (TRIM)-containing proteins play crucial roles in regulating critical cellular biological processes, including differentiation, proliferation, apo- ptosis, autophagy, signal transduction, innate immunity, and tumorigenesis [24-29]. TRIM21 is implicated in the regulation of differentiation in immune cells, senescence and proliferation in osteosarcoma cells, osteoblast differentiation of osteosarcoma cells, and osteogenic differentiation of bone mesenchymal stem cells (BMSCs) [30-36]. TRIM16 promotes osteogenic differentiation of human periodontal ligament stem cells by regulating RUNX2 stability [37]. TRIM38 is a key factor in the regulation of bone remodeling by controlling osteoblast and osteoclast differentiation [38]. TRIM65 plays a role in regulating various biological processes, such as cell proliferation and migration [39-42], cell growth [43], apoptosis [44, 45], autophagy [45, 46], and inflammation [47-49]. Osteoblasts play a crucial role in OP development and progression. However, whether TRIM65 participates in osteoblastogenesis and contributes to OP remains unclear.
In our study, we observed that TRIM65 expression significantly increased during the osteogenic differenti- ation of BMSCs and a mouse preosteoblastic cell line (MC3T3-E1 cells). Moreover, we observed that TRIM65 overexpression promoted osteogenic differentiation of MC3T3-E1 cells compared to the control group. In contrast, the loss of TRIM65 inhibited the osteogenic differentiation of BMSCs and MC3T3-E1 cells as compared to the control group. Furthermore, the PI3K/AKT signaling pathway was activated post-TRIM65 knockout during the osteogenic differentiation of BMSCs and MC3T3-E1 cells. These findings suggest that TRIM65 plays a role in OP regulation by affecting osteogenic differentiation.
2. MATERIALS AND METHODS
2.1. Cell Culture and Differentiation
Primary mouse BMSC extracts and cultures were prepared as previously described [50], and the study was approved by the Institutional Ethical Board of the University of South China. Three six-week-old male mice (C57BL/6J mice) were sacrificed by cervical dislocation and soaked in 75% alcohol for 3–5 min. The four limb bones were aseptically extracted from the mice. Bone marrow was obtained from long bones of mice with α modified Eagle medium (α-MEM; AW-M011, Abiowell, China) using a sterile syringe to rinse the bone marrow cavity. The bone marrow fluid was centrifuged for 10 min at 1500 rpm. The supernatant was removed. We added α-MEM complete medium (AW-MC023, Abiowell, China) containing 10% fetal bovine serum (FBS) and 500 U/mL penicillin/streptomycin (P/S) and cultured at 37 °C and 5% CO2 for 48–96 h. Two to three passages of BMSCs were used for all experiments.
The MC3T3-E1 cells were purchased from Abiowell Biological Technology Co., Ltd. (AW-CNM401, China) and cultured with α-MEM complete medium. For induction of differentiation, BMSCs or MC3T3‐E1 cells were seeded into a 6‐well plate at a density of 5×105 cells/well. On reaching 80–90% confluence, the medium was discarded, changed with osteogenic differentiation medium (OriCell, MUXMT-90021), and cultured at 37 °C with 5% CO2 for 14 days. The culture medium was replaced every 2 or 3 days. Alizarin red S (ARS), alkaline phosphatase (ALP), immunofluorescence staining, and western blotting were conducted to examine osteoblastic differentiation.
2.2. Immunofluorescence Staining
The cells were seeded on coverslips in 48-well plates and induced with osteogenic differentiation medium as described above. The cells were then fixed for 30 min, permeabilized with 0.3% Triton X‐100 for 20 min, and blocked with 10% FBS for 1 h. The cells were incubated with the primary antibodies (TRIM65, Biorbyt; COL1A1, Proteintech; RUNX2, Proteintech; Osteocalcin, Pro- teintech) overnight at 4 °C and with secondary antibodies (Alexa Fluor® 488 or 594) in the dark for 1 h at 37 °C, followed by 4′,6‐diamidino‐2‐phenylindole (DAPI) for 5 min. The cells were observed under a fluorescent inverted microscope and photographed.
2.3. Lentivirus Transfection and Stable Lines Screening
The overexpressing TRIM65 lentivirus plasmid, mouse TRIM65 gene-targeting siRNA, and their control plasmid were purchased from Jikai Biological Company (Shanghai, China). MC3T3‐E1 cells were infected with four plasmids (TRIM65, EV, shTRIM65, and NC). After transfection for 72 h, the cells were treated with puromycin (1 μg/mL) for 7 days. Viable cells were used for subsequent experiments. Infection efficiency was observed using fluorescence microscopy, real-time fluorescence quantitative poly- merase chain reaction (qRT-PCR), and western blotting.
2.4. Western Blotting
The total protein was extracted by using RIPA lysis buffer (CWBIO, CW2333S, China) containing 1% phenyl- methylsulfonyl fluoride (PMSF; Beyotime, ST507, China) and 1% phosphatase inhibitors (CWBIO; CW2383S, China). Protein concentration was determined by using a bicinchoninic acid protein assay kit (CWBIO, CW0014S, China). Protein samples were diluted with 5×SDS-PAGE loading buffer (CWBIO, CW0028S, China) at 95 °C for 8 min. Then, 50μg proteins were separated using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins were then transferred to poly- vinylidene difluoride membranes (PVDF; Millipore, IPVH00010, MA). They were blocked with 5% nonfat milk for 2–4 h at room temperature and incubated with primary antibodies against TRIM65 (1:1000; Biorbyt), COL1A1 (1:500; Proteintech), RUNX2 (1:1000; Proteintech), OCN (1:2000; Proteintech), phospho-PI3K (1:500; Proteintech), PI3K (1:1,000; Proteintech), phospho-AKT (1:500; Proteintech), AKT (1:500; Proteintech), and GAPDH (1:10000; Proteintech) at 4 °C overnight. The membranes were incubated with the corresponding secondary antibodies (1:5000; Proteintech) on a shaker for 2 h at room temperature. Immunodetection was conducted with an Omni-ECL™ ultra-sensitive chemiluminescence test kit (Epzyme, SQ201, China) and Image Quant LAS 4000. Protein bands were quantitatively analyzed using the ImageJ software.
2.5. q-PCR
The total RNA was extracted using the TRIzol Reagent (CWBIO, CW0580S, China). cDNA was synthesized using the SuperStar miRNA first-strand cDNA synthesis kit (by tailing A) (CWBIO, CW2151M, China). PCR was performed with a SYBR PCR kit (CWBIO, CW3360M, China) on a StepOne Real‐time PCR System (ABI Company, USA). We used reduced glyceraldehyde-phosphate dehydrogenase (GAPDH) as the endogenous control. The 2−ΔΔCt was used to evaluate the relative mRNA expression levels. The primers sequences of TRIM65 and GAPDH are listed as follows [48]: TRIM65-Forward: 5′-AAGAGAAGAGCCTCC- CCAAG-3′, TRIM65-Reverse: 5′-GGTCTCTGGGTCAAA- GGTCA-3′; GAPDH-Forward: 5′-ACCCAGAAGACTGTGG- ATGG-3′, GAPDH-Reverse: 5′-ACACATTGGGGGTAGG- AACA-3′.
2.6. ALP Staining
Cells were seeded into 12-well plates at a density of 5 × 104 cells/well and induced with osteogenic differenti- ation medium as described earlier. An ALP staining reagent (Solarbio, G1480, China) was used to determine ALP activity according to the instructions of the manufacturer. Stained images were observed and captured in five random fields using a phase-contrast microscope (Nikon, Japan).
2.7. ARS Staining
The cells induced in 12-well plates were cleaned and fixed for 20–30 min. Then, the cells were stained with 0.1% alizarin red staining solution (OriCell, ALIR-10001) for 20 min. After cleaning thrice, calcium nodule formation was observed using a phase-contrast microscope (Nikon, Tokyo, Japan). The images were captured in five random fields.
2.8. RNA-sequencing Analysis
Transcriptome sequencing was conducted by Origin- Gene Bio-Pharm Technology Co., Ltd. The experimental process was as follows: MC3T3-E1 cells from shNC and shTRIM65 groups were collected using TRIzol Reagent (CWBIO, CW0580S, China). After extracting the total RNA from the sample, rRNA was cleared using an rRNA removal kit, mRNA was enriched, and double-stranded cDNA was synthesized. The double-ended cDNA was repaired, a splice was added, and cDNA libraries were constructed by PCR amplification. RNA-seq was conducted using an Illumina high-throughput sequencing platform.
2.9. Statistical Analysis
All results are represented as mean ± standard deviation (x ± SD). The students' t-test was used to make pairwise comparisons. One-way analysis of variance (ANOVA) was used for comparisons among more than two groups. All experiments were conducted thrice indepen- dently. All data analyses were conducted using the ImageJ and GraphPad Prism 7.0 software. A p-value < 0.05 was considered to be statistically significant.
3. RESULTS
3.1. TRIM65 Expression Increased during Osteogenic Differentiation
To investigate whether TRIM65 is involved in osteoblast differentiation, western blotting and immuno- fluorescence staining were conducted to detect TRIM65 expression after the cells' induction with osteogenic differentiation medium on days 0, 7, and 14 (Fig. 1A-1G). We found that TRIM65 predominantly existed in the cytoplasm of MC3T3‐E1 cells and BMSCs. The results demonstrated that TRIM65 levels were significantly up-regulated in the induced cells (Fig. 1H–1M). This suggests that TRIM65 participates in the osteogenic differentiation of BMSCs and MC3T3-E1 cells.
3.2. Effect of TRIM65 Overexpression on Osteogenic Differentiation
To further explore the effect of TRIM65 on osteogenic differentiation, we constructed an overexpressing plasmid of TRIM65 (LV-TRIM65) and its control (LV-empty vector, EV) vector using a lentivirus. MC3T3-E1 cells were infected with an overexpressing TRIM65 lentiviral plasmid (LV-TRIM65) and its control (EV). At 72 h post-infection, puromycin was used to screen for positive cells. Transfection efficiency was assessed by fluorescence microscopy and western blotting. As depicted in Fig. (2A–2C), TRIM65 expression increased 3.8 folds in LV-TRIM65-transfected MC3T3-E1 cells compared to that in EV cells. Furthermore, we evaluated the effect of TRIM65 on the osteogenic differentiation and mineralization of MC3T3-E1 cells by measuring the protein expression of the osteogenesis-related genes COL1A1, RUNX2, and OCN using western blotting and immunofluorescence staining. The results demonstrated that TRIM65 overexpression significantly increased COL1A1, RUNX2, and OCN (Fig. 2D-2F). The effect of TRIM65 on ALP activity, another important phenotypic marker of differentiation, was determined by ALP staining. The results demonstrated that TRIM65 significantly increased ALP activity compared to that in the EV group (Fig. 2G). Additionally, the effect of TRIM65 on calcified bone nodules was determined by ARS staining 14 days after the osteogenic differentiation of MC3T3-E1 cells. As depicted in Fig. (2H), more calcium bone nodules were found in TRIM65-overexpressed MC3T3-E1 cells than in the EV group. This indicates that TRIM65 overexpression promotes osteogenic differenti- ation of MC3T3-E1 cells.
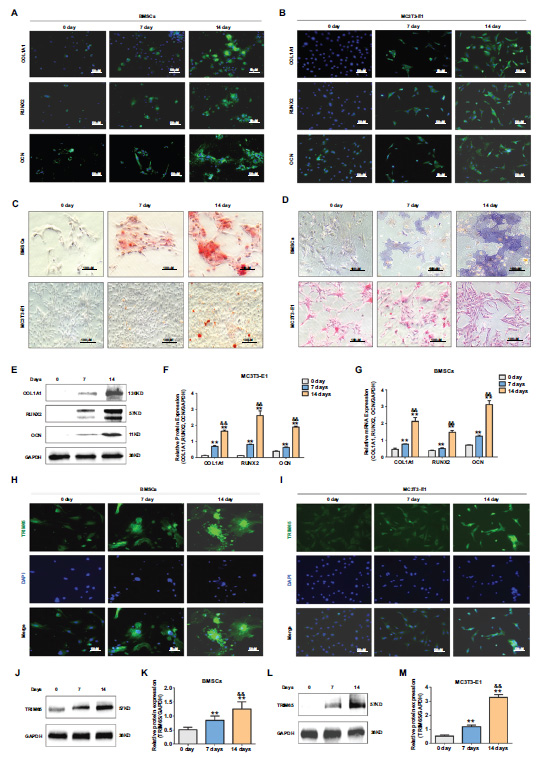
TRIM65 expression increased during osteoblast differentiation of BMSCs and MC3T3-E1 cells. (A) The expression of COL1A1, RUNX2, and OCN was detected by Immunofluorescence (IF) staining during the osteoblast differentiation of BMSCs on days 0, 7, and 14. (B) MC3T3-E1 cells were cultured in an osteogenic medium on days 0, 7, and 14. IF staining was used to detect COL1A1, RUNX2, and OCN expression. (C) ARS was used to detect the calcium nodule formation during the osteoblast differentiation of BMSCs and MC3T3-E1 cells on days 0, 7, and 14. (D) ALP staining was used to detect ALP activity during the osteoblast differentiation of BMSCs and MC3T3-E1 cells on days 0, 7, and 14. (E) Western blotting and quantitative analysis (F) were used to detect the expression of COL1A1, RUNX2, and OCN during the osteoblast differentiation of MC3T3-E1 cells on days 0, 7, and 14. (G) q-PCR was used to detect the expression of COL1A1, RUNX2, and OCN during the osteoblast differentiation of BMSCs on days 0, 7, and 14. (H) TRIM65 expression was detected during the osteoblast differentiation of BMSCs on days 0, 7, and 14 using an IF staining assay. (I) MC3T3-E1 cells were cultured in an osteogenic medium for 0, 7, and 14 days, and IF staining was used to detect TRIM65 expression during the osteoblast differentiation of BMSCs on days 0, 7, and 14. (J) Western blotting and quantitative analysis (K) were used to detect TRIM65 expression during the osteoblast differentiation of BMSCs cells on days 0, 7, and 14. (L) Western blotting and quantitative analysis (M) were used to detect TRIM65 expression during the osteoblast differentiation of MC3T3-E1 cells on days 0, 7, and 14. Error bars are presented as the mean ± SD, and comparisons were performed using one‐way ANOVA. **P < 0.01 vs. 0 days; &&P < 0.01 vs. 7 days, n=3 in each group.
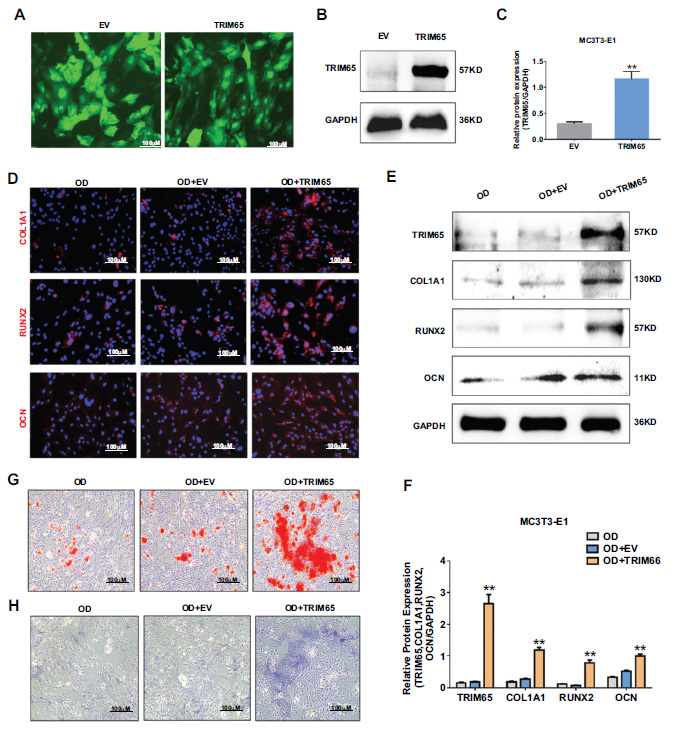
Effect of TRIM65 on the osteoblast differentiation of MC3T3-E1 cells. (A) MC3T3-E1 cells were infected with overexpressed TRIM65 lentivirus plasmid (LV-TRIM65) and its control (EV). The fluorescence intensities were observed using an inverted fluorescence microscope. (B) Western blotting and quantitative analysis (C) were used to detect the expression of TRIM65 in MC3T3-E1 cells after the cells were transfected with LV-TRIM65. (D) MC3T3-E1 cells overexpressing with TRIM65 were cultured in osteogenic medium for 14 days, and immunofluorescence staining was used to detect COL1A1, RUNX2, and OCN expression. (E) Western blotting and quantitative analysis (F) were used to detect the expression of COL1A1, RUNX2, and OCN during the osteoblast differentiation of TRIM65-overexpressed MC3T3-E1 cells at 14 days. (G) ARS was used to detect the calcium nodule formation during the osteoblast differentiation of TRIM65-overexpressed MC3T3-E1 cells at 14 days. (H) ALP staining was used to detect the ALP activity during the osteoblast differentiation of TRIM65-overexpressed MC3T3-E1 cells at 14 days. Error bars are presented as the mean ± SD, and comparisons were performed using one‐way ANOVA. **P < 0.01 vs. EV group, n=3 in each group.
3.3. Effect of TRIM65 Knockdown on Osteogenic Differentiation
To explore the role of TRIM65 in osteogenic differentiation, we inhibited TRIM65 expression by transfecting with shRNA-TRIM65 lentivirus plasmid (shTRIM65) and its control (shNC). Transfection efficiency was determined by western blotting. The results demonstrated that the protein expression of TRIM65 was significantly reduced (Fig. 3A and 3B). In addition, we evaluated the effect of TRIM65 knockdown on the osteogenic differentiation and mineralization of MC3T3-E1 cells. We measured the protein expression of COL1A1, RUNX2, and OCN with immunofluorescence staining and western blotting. The results revealed that TRIM65 deficiency significantly reduced COL1A1, RUNX2, and OCN expression (Fig. 2C-2E). Moreover, the ALP activity and calcified bone nodules were significantly decreased after TRIM65 was knocked down (Fig. 2F and 2G). This suggests that TRIM65 deletion inhibits osteogenic differentiation of MC3T3-E1 cells.
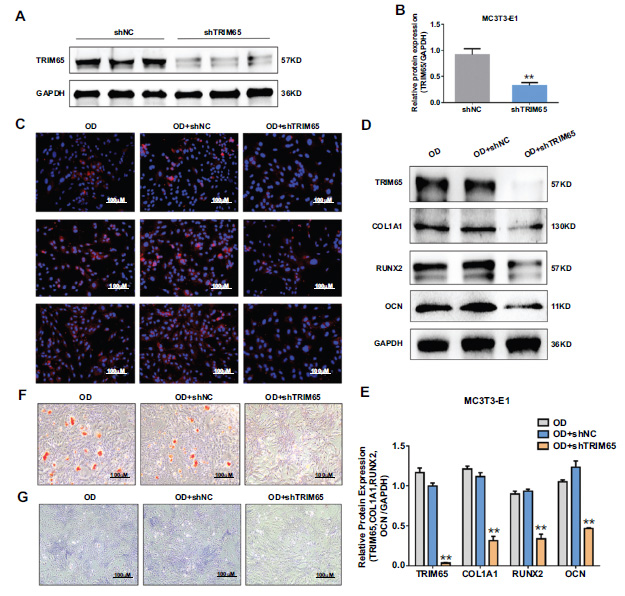
Effect of TRIM65 knockdown on osteoblast differentiation of MC3T3-E1 cells. (A) MC3T3-E1 cells were infected with shRNA-TRIM65 lentivirus plasmid (shTRIM65) and its control (shNC). TRIM65 protein expression was determined by using western blotting analysis. (B) The gray value of Figure A is quantitatively analyzed by using Image J and the GraphPad Prism 7.0 software. The error bar was presented as the mean ± SD, and comparisons were performed using student t-test. **P < 0.01 vs. shNC, n=3. (C) MC3T3-E1 cells were transfected with shTRIM65 and cultured in osteogenic medium for 14 days, and immunofluorescence staining was used to detect COL1A1, RUNX2, and OCN expression. (D) Western blotting and quantitative analysis (E) were used to detect the expression of COL1A1, RUNX2, and OCN during the osteoblast differentiation of TRIM65-knockdown MC3T3-E1 cells at 14 days. (F) ARS was used to detect the calcium nodule formation during the osteoblast differentiation of TRIM65-knockdown MC3T3-E1 cells at 14 days. (G) ALP staining was used to detect the ALP activity during the osteoblast differentiation of TRIM65-knockdown MC3T3-E1 cells at 14 days. Error bars are presented as the mean ± SD, and comparisons were performed using one‐way ANOVA. **P < 0.01 vs. OD, n=3 in each group, OD: osteogenic differentiation.
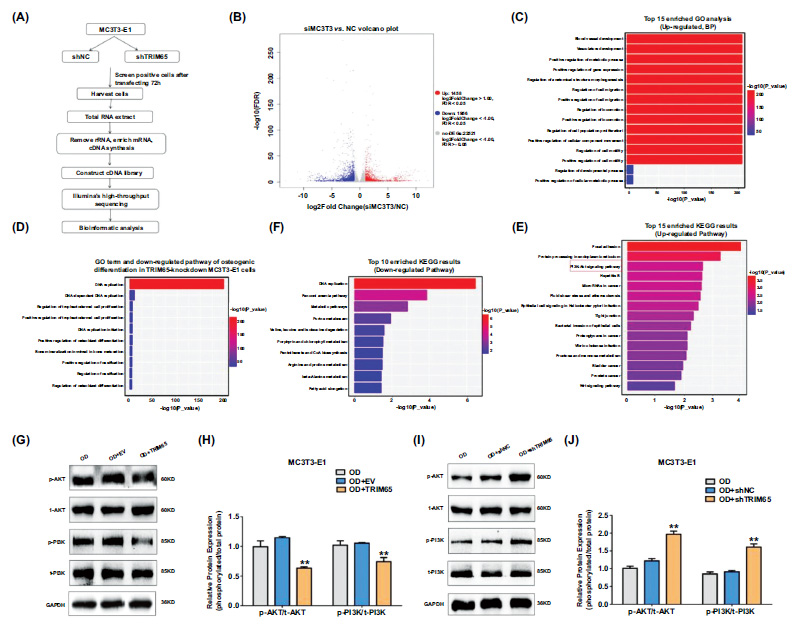
Depletion of TRIM65 inhibits osteogenic differentiation by regulation of PI3K/AKT pathway (A) Schematic diagram showing RNA-seq analysis was performed on shNC and shTRIM65 groups for identification of DEGs in MC3T3-E1 cells. (B) The volcano map shows DEGs in shNC and shTRIM65 transfected MC3T3-E1 cells. (C) GO enrichment analysis of the top 15 up-regulated DEPs in shNC and shTRIM65 transfected MC3T3-E1 cells. (D) GO enrichment analysis of the top 10 down-regulated pathways of osteogenic differentiation in shNC and shTRIM65 transfected MC3T3-E1 cells. (E) KEGG enrichment analysis of the top 15 up-regulated DEPs in shNC and shTRIM65 transfected MC3T3-E1 cells. (F) KEGG analysis of the top 10 down-regulated pathways in shNC and shTRIM65 transfected MC3T3-E1 cells. (G) Western blotting and quantitative analysis (H) were used to detect the ratio of phosphorylated proteins and total protein expression of PI3K and AKT during the osteoblast differentiation of TRIM65-overexpressed MC3T3-E1 cells at 14 days. (I) Western blotting and quantitative analysis (J) were used to detect the ratio of phosphorylated proteins and total protein expression of PI3K and AKT during the osteoblast differentiation of TRIM65-knockdown MC3T3-E1 cells at 14 days. Error bars are presented as the mean ± SD, and comparisons were performed using two‐way ANOVA. **P < 0.01 vs. OD, n=3, OD:osteogenic differentiation.
3.4. Involvement of PI3K/AKT Pathway in TRIM65-regulated Osteogenic Differentiation
To reveal the mechanisms underlying the role of TRIM65 in the osteogenic differentiation of MC3T3-E1 cells, RNA-seq was performed on shNC and shTRIM65 groups (Fig. 4A). A total of 25,935 genes were identified in the MC3T3-E1 cells transfected with shTRIM65 compared to the shNC group. Among the identified genes, 1,458 were up-regulated, and 1,956 were down-regulated, as depicted in the volcano map (Fig. 4B). To further clarify the effect of shTRIM65 on the biological functions of MC3T3-E1 cells, we selected the top 15 enriched Gene Ontology (GO) terms for biological processes (BP) to facilitate the analysis based on the up-regulated differentially expressed genes (DEGs). The data depicted that up-regulated DEGs were predominantly involved in blood vessel and vasculature development, ossification, gene expression, metabolic processes, cell migration and
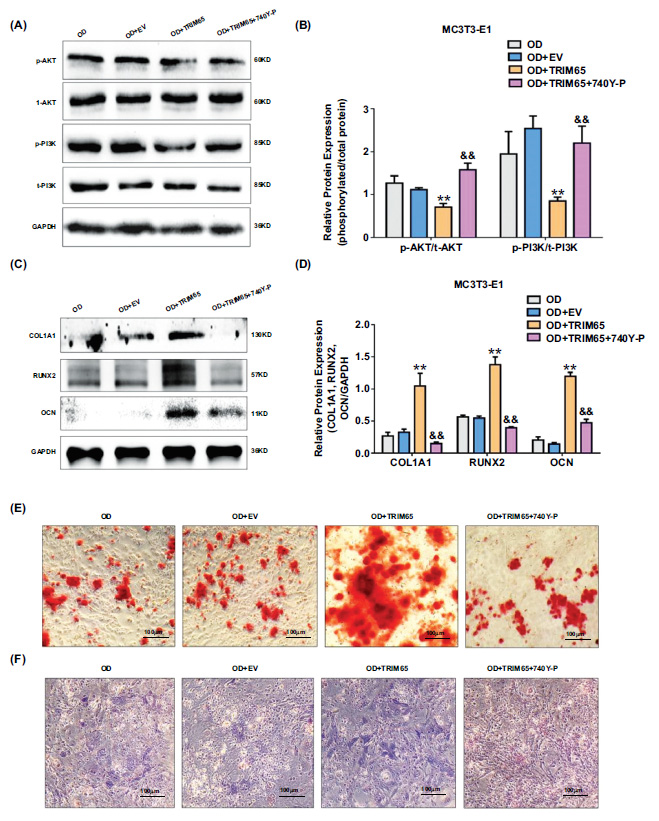
TRIM65 promotes osteogenic differentiation of MC3T3-E1 cells by modulating the PI3K/AKT pathway. (A) TRIM65-overexpressed MC3T3-E1 cells were treated with 740Y-P. The expressions of phosphorylated PI3K and AKT proteins were detected by using western blotting analysis. (B) Quantitative analysis was used to detect the ratio of phosphorylated proteins and total protein expression of PI3K and AKT in TRIM65-overexpressed MC3T3-E1 cells after treatment with 740Y-P. Error bars are presented as the mean ± SD, and comparisons were performed using two‐way ANOVA. **P < 0.01 vs. OD, &&P < 0.01 vs. OD+TRIM65, n=3 in each group. (C) Western blotting and (D) quantitative analysis were used to detect COL1A1, RUNX2, and OCN expression. Error bars are presented as the mean ± SD, and comparisons were performed using two‐way ANOVA. **P < 0.01 vs. OD, &&P < 0.01 vs. OD+TRIM65, n=3 in each group. (E) ARS was used to detect the calcium nodule formation during the osteoblast differentiation of TRIM65-overexpressed MC3T3-E1 cells after treatment with 740Y-P. Scale bar=100μm. (F) ALP staining was used to detect the ALP activity during the osteoblast differentiation of TRIM65-overexpressed MC3T3-E1 cells after treatment with 740Y-P. Scale bar=100μm. OD: osteogenic differentiation.
proliferation, cell movement, and motility (Fig. 4C). To further screen the key genes involved in OB differentiation from these DEGs, we selected the top 10 enriched GO terms of BP to facilitate the analysis according to the down-regulated DEGs. This demonstrated that the major down-regulated differential genes post TRIM65 knock- down in the MC3T3-E1 cells were involved in the regulation of DNA replication, cell proliferation, osteoblast differentiation, ossification and bone maturation, and bone calcification (Fig. 4D). Kyoto Encyclopedia of Genes and Genomes Pathway (KEGG) analysis revealed that the up-regulated DEGs were involved in the PI3K/AKT pathway (Fig. 4E). The top 10 down-regulated DEGs were predominantly enriched in the regulation of DNA replication and metabolic pathways (Fig. 4F). To verify whether the PI3K/AKT pathway is involved in TRIM65-regulated osteogenic differentiation, we examined the expression of phosphorylated proteins and total protein expression of PI3K and AKT in TRIM65-overexpressed or TRIM65-knockdown MC3T3-E1 cells using western blotting analysis. The expression of phosphorylated PI3K and AKT was significantly down-regulated by TRIM65 overexpression in the MC3T3-E1 cells (Fig. 4G and 4H). In contrast, p-PI3K and p-AKT levels were significantly up-regulated in the TRIM65-knockdown cells (Fig. 4I and 4J). This was consistent with the results of KEGG analysis, revealing that the PI3K/AKT pathway is associated with TRIM65-regulated osteogenic differentiation.
3.5. TRIM65 Promotes Osteogenic Differentiation of MC3T3-E1 Cells by Modulating the PI3K/AKT Pathway
To explore whether TRIM65 promotes osteogenic differentiation of MC3T3-E1 cells by regulating the PI3K/AKT pathway, we examined the protein expression of COL1A1, RUNX2, OCN, and the ALP activity, and calcified bone nodules in TRIM65-overexpressed MC3T3-E1 cells after treatment with 740Y-P, a PI3K/AKT agonist. We found that the expressions of phosphorylated PI3K and AKT proteins were up-regulated (Fig. 5A and 5B), and COL1A1, RUNX2, and OCN levels were down-regulated (Fig. 5C and 5D). Furthermore, the ALP activity and calcified bone nodules were inhibited (Fig. 5E and 5F) after TRIM65-overexpressed MC3T3-E1 cells treatment with 740Y-P. These results revealed that TRIM65 promotes osteogenic differentiation of MC3T3-E1 cells by regulating the PI3K/AKT pathway.
4. DISCUSSION
OP is a widespread chronic disease characterized by reduced bone mass or low bone density, which can reduce bone strength and lead to fractures [51-55]. Osteoporotic fractures are gaining increasing attention because of their high incidence rates and lack of satisfactory treatment for OP. Osteoblasts, which are the sole bone-forming cells, are responsible for the formation of bones and the maintenance of bone mass [56-60]. Osteoblast differentiation is crucial for maintaining bone strength. Herein, we aimed to identify a novel target for OP treatment that regulates osteoblast differentiation.
An increasing number of studies have emphasized the importance of TRIM65 in tumorigenesis [61], inflam- matory response [62, 63], and cell proliferation and migration [39]. Osteogenic differentiation is a key process in bone formation [60]. Dysfunction of osteoblastic bone formation is associated with the occurrence and progression of OP. In this study, we found that TRIM65 was up-regulated during osteogenic differentiation of BMSCs and MC3T3-E1 cells. We discovered for the first time that TRIM65 overexpression promotes osteoblastic bone formation by elevating the differentiation of MC3T3-E1 cells and increasing the formation of osteoblasts. In contrast, TRIM65 depletion inhibited osteoblast bone formation by preventing osteoblast osteogenic differenti- ation and attenuating ALP generation. Therefore, TRIM65 may play a crucial role in osteogenic differentiation.
PI3K/AKT signaling plays a crucial role in cell proliferation and differentiation [64-67]. This pathway is crucial for osteoblast differentiation, bone formation, and bone regeneration [68-70]. Our cooperative team, Zhou et al., previously reported that TRIM65 plays a critical role in PI3K/AKT/mTOR signaling during vascular smooth muscle cells (VSMCs) phenotypic transformation and athero- genesis [39]. Herein, bioinformatics analysis of RNA sequences in the shTRIM65- and shNC-transfected MC3T3-E1 cells revealed that TRIM65 majorly affected PI3K/AKT signaling. Moreover, our results indicated that TRIM65 overexpression in the MC3T3-E1 cells strongly inhibited the expression of phosphorylated PI3K and AKT. Conversely, TRIM65 knockdown in the MC3T3-E1 cells significantly increased the expression ratios of p-PI3K/PI3K and p-AKT/AKT. Therefore, TRIM65-modulated osteoblast differentiation was accomplished by regulating the PI3K/AKT pathway. However, this study was conducted in vitro. Thus, we will use TRIM65 knockout animal models to further investigate the effects of TRIM65 on osteoblast differentiation and OP.
CONCLUSION
Our experiments revealed that TRIM65 was elevated during the osteoblast differentiation of BMSCs and MC3T3-E1 cells. We further demonstrated that TRIM65 overexpression increased MC3T3-E1 cell differentiation, promoted osteogenic bone formation, and enhanced ALP levels, whereas TRIM65 deletion exhibited the opposite results. Moreover, the PI3K/AKT pathway was considered and determined to be triggered after TRIM65 deletion in MC3T3-E1 cells. This study provides a new intervention target for TRIM65 for the prevention and treatment of OP.
AUTHORS’ CONTRIBUTION
J. X., H.-P. L. and T.-T. Z. conceived the idea and designed the experiments. Z.-X. Z. contributed to preparing the manuscript for publication. Y.-F. L. analyzed the data. X. X. and A.-Y. Liang took part in the data collection. M. W, Y.-J. L. and J. X. contributed to data analysis and interpretation.
LIST OF ABBREVIATIONS
| OP | = Osteoporosis |
| BMSCs | = Bone Mesenchymal Stem Cells |
| FBS | = Fetal Bovine Serum |
| ARS | = Alizarin red S |
| qRT-PCR | = Quantitative Polymerase Chain Reaction |
| SDS-PAGE | = Sodium Dodecyl Sulfate-polyacrylamide Gel Electrophoresis |
| GO | = Gene Ontology |
| BP | = Biological Processes |
| DEGs | = Differentially Expressed Genes |
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This study was approved by the Institutional Ethical Board of the University of South China, China, (approval No.vsc20180705XS036).
HUMAN AND ANIMAL RIGHTS
This study adhered to internationally accepted standards for animal research, following the 3Rs principle. The ARRIVE guidelines were employed for reporting experiments involving live animals, promoting ethical research practices.
AVAILABILITY OF DATA AND MATERIAL
The data supporting the findings of the article is available in the (Zenodo Repository) at (Open Medicine Journal), reference number (BMS-MEDJ-2024-21).
FUNDING
This work was supported by the Hunan Provincial Department of Education, China key project (23A0545 to XJ), the General Guidence Project of Scientific Research Program of Hunan Provincial Health Commission (202302069287 to ZXZ), the Scientific Research Project of Hunan Provincial Department of Education (22B0408 to ZXZ).


