RESEARCH ARTICLE
CLEC4A Expression as a Prognostic Biomarker and Immunoregulator in Lung Adenocarcinoma: Insights from Immune Cell Infiltration
Huiyun Ma1, 2, #, Gujie Wu1, #, Hongyu Chen1, #, Qin Hu1, Zhouwei Zhang1, Fei Wang1, *, Qun Xue1, *
Article Information
Identifiers and Pagination:
Year: 2024Volume: 11
E-location ID: e18742203270381
Publisher ID: e18742203270381
DOI: 10.2174/0118742203270381240209060006
Article History:
Received Date: 25/09/2023Revision Received Date: 15/02/2024
Acceptance Date: 15/02/2024
Electronic publication date: 18/4/2024
Collection year: 2024

open-access license: This is an open access article distributed under the terms of the Creative Commons Attribution 4.0 International Public License (CC-BY 4.0), a copy of which is available at: https://creativecommons.org/licenses/by/4.0/legalcode. This license permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Abstract
Background
CLEC4A (C-type lectin domain family 4 member A), a member of the C-type (Ca2+-dependent) lectin (CLEC) receptor, is an immunosuppressant of dendritic cells (DCs) and plays an important role in innate and adaptive immunity, however, its role in lung adenocarcinoma (LUAD) and the potential for immunotherapy remains to be investigated.
Methods
To achieve our objectives, we conducted a comprehensive analysis of CLEC4A expression and its correlation with clinical factors in LUAD. We utilized publicly available datasets, such as The Cancer Genome Atlas (TCGA) and other relevant resources, to gather gene expression and clinical data from LUAD patients. Furthermore, we investigated the association of CLEC4A expression levels with clinical pathological staging and prognosis of lung adenocarcinoma. The TIMER database was utilized to analyze immune cell infiltration, while the TISIDB database provided insights into lymphocyte infiltration and immune regulatory factors.
Results
Our analysis revealed a significant correlation between poor prognosis and low CLEC4A expression in LUAD patients. Reduced expression of CLEC4A was associated with adverse clinical factors, indicating its potential as a prognostic biomarker in LUAD. Moreover, we observed a noteworthy relationship between CLEC4A expression and immune cell infiltration. Increased CLEC4A expression was correlated with higher infiltration levels of CD8+ T cells, CD4+ T cells, dendritic cells (DC), and B cells within the tumor microenvironment. This indicates an immunoregulatory role for CLEC4A in modulating immune responses against LUAD. Additionally, our analysis highlighted a positive correlation between CLEC4A expression and the presence of lymphocytes, further emphasizing its potential importance in tumor immunity. Furthermore, the investigation of immune-related factors indicated a potential involvement of CLEC4A in immune regulation within the tumor microenvironment.
Conclusion
This study provides valuable insights into the expression, prognosis, and potential immunotherapeutic role of CLEC4A in lung adenocarcinoma (LUAD). The identified correlations between CLEC4A expression and clinical characteristics, immune cell infiltration, and lymphocyte infiltration highlight the significance of CLEC4A as a potential biomarker and therapeutic target for LUAD. Further research is warranted to elucidate the underlying mechanisms and capitalize on the therapeutic potential of targeting CLEC4A in LUAD. These efforts could contribute to improving patient outcomes and prognosis in LUAD.
1. INTRODUCTION
Lung cancer is a global health burden characterized by its high morbidity and mortality rates [1]. It is categorized into two main types: small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), with NSCLC accounting for approximately 85% of cases [2]. NSCLC can be further divided into subtypes, including squamous cell carcinoma, adenocarcinoma, and large cell carcinoma. Adenocarcinoma is the most prevalent subtype, repre- senting about 40% of all lung cancer cases and is known for its aggressive nature and poor prognosis [3]. Although early surgical resection of lung adenocarcinoma (LUAD) offers a favorable prognosis, the majority of patients are unfortunately diagnosed at an advanced stage, hindering the effectiveness of treatment [4]. In recent years, advancements in driver gene-targeted therapy and immunotherapy have revolutionized traditional radio- therapy approaches. Despite these advancements, the 5-year survival rate for LUAD patients remains dis- hearteningly low, standing at only 15% [5, 6]. This highlights the critical need to identify novel markers and therapeutic targets for LUAD to improve patient prognosis.
CLEC4A, a C-type lectin receptor (CLR), is expressed by various immune cells, particularly dendritic cells (DCs) [7]. The cytoplasmic tail of CLEC4A contains an immune receptor tyrosine-based inhibitory signaling motif (ITIM). It plays a crucial role in regulating immune receptor tyrosine-activated motif (ITAM)-mediated activation of Src homologous phosphatase (SHP), an important signaling molecule associated with inhibiting DC maturation [8]. CLEC4A expression is downregulated by signals inducing DC maturation, such as tumor necrosis factor-alpha (TNF-α), lipopolysaccharide (LPS), and CD40 ligand stimulation. DCs possess the ability to capture and present antigens, and CLEC4A acts as an antigen-delivering receptor, activating specific CD8+ T cell immune responses [9]. Given these characteristics, CLEC4A holds promise as an immunotherapeutic target, and its role as a biomarker for LUAD deserves exploration.
In our comprehensive medical study, we analyzed the expression and prognosis of CLEC4A in lung adenocarcinoma (LUAD) using the COX regression method and survival analysis. Our findings revealed a strong correlation between poor prognosis and various clinical factors associated with LUAD. This significant correlation highlights the importance of CLEC4A in evaluating the overall prognosis of patients with LUAD. In addition, we further explored the relationship between CLEC4A and the tumor microenvironment, specifically investigating the association between CLEC4A and infiltrating immune cells (particularly lymphocytes) as well as immune-related factors. This aspect of our study sheds light on the potential mechanisms underlying CLEC4A's role in tumor immunity. By uncovering the connection between CLEC4A and immune cells, we have paved the way for future research aimed at targeting CLEC4A in the context of tumor immunity. By thoroughly analyzing the expression and prognostic implications of CLEC4A in LUAD, we have contributed valuable insights into understanding the clinical relevance of this particular gene in lung adenocarcinoma. Furthermore, our investigation into the relationship between CLEC4A and infiltrating immune cells and immune-related factors expands the current knowledge base and provides a foundation for further exploration of CLEC4A as a potential therapeutic target in tumor immunity.
2. MATERIALS AND METHODS
2.1. Data Collection
To investigate the RNA expression profiles and clinical characteristics of patients with lung adenocarcinoma (LUAD), our study utilized data from the TCGA database (https://portal.gdc.cancer.gov/). Utilizing this compre- hensive resource, we obtained RNA-seq data as well as corresponding clinical information for a total of 535 LUAD patients. In order to establish a reliable baseline for comparison, we also collected RNA-seq data for 59 normal lung tissues. By obtaining a substantial dataset comprising both cancerous and normal lung tissue, we were able to perform robust analysis and draw meaningful conclusions. The inclusion of such a diverse sample set provides a comprehensive overview of the RNA expression profiles specific to LUAD, enabling us to make accurate assessments of the disease at the molecular level.
2.2. Cell Culture
For our experimental analysis, we obtained various LUAD cell lines (H1975, H1299, H23) and a normal lung cell line (16HBE) from Procell. These cell lines were cultured in sterile culture dishes using RPMI-1640 medium supplemented with 10% fetal bovine serum. The cells were incubated at 37°C in a 5% CO2 environment with saturated humidity, maintaining optimal conditions for their growth and proliferation.
2.3. RNA Isolation and qRT-PCR
To investigate the RNA expression levels in LUAD cells, we isolated and extracted RNA using the TRIzol reagent (Invitrogen). The concentration of RNA was determined, and subsequently, reverse transcription into cDNA was performed using the Reverse Transcription System kit. Finally, we conducted RT-qPCR reactions to quantify the expression levels of the target genes. The primer sequences used for this analysis were as follows: CLEC4A: Forward 5'-ATGAACAGGTGGTTGGATTGG-3', Reverse 5'-CCAGTACAGGCTCTGGCACAT-3'; ACTIN: Forward 5'-ACTTAGTTG CGTTACACCCTTTC-3', Reverse 5'-TCACCTTCACCGTTCCAGTTT-3'.
2.4. Immunological Infiltration Analysis
To gain insights into the immune infiltrate composition within the tumor microenvironment, we referred to Bindea G's study [10] to obtain marker genes specific to 24 different immune cells. Exploring the infiltration patterns of immune cells in both high and low CLEC4A expression groups, we employed the ssGSEA algorithm and conducted a statistical analysis using the Wilcoxon rank sum test. Furthermore, we investigated the correlation between CLEC4A and the aforementioned 24 immune cell populations using the Spearman correlation method.
2.5. Differential Expression Analysis
The RNAseq data in HTSeq- FPKM format was converted to TPM format, and log2 was transformed. The Wilcoxon rank sum test was used to analyze the expression of CLEC4A in pan-cancer. The results of the differences between paired and unpaired samples for LUAD in TCGA were then presented using grouped comparison plots.
2.6. Validation Analysis
The expression profiles and clinical information of GSE75037 and GSE68465 were downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds) and used to validate the differential expression and prognostic value of CLEC4A in LUAD.
2.7. TISIDB Database
The TISIDB database (http://cis.hku.hk/TISIDB) integrates multiple data sources in tumor immunology and contains 988 genes associated with anti-tumour immunity to study tumour-immune interactions. Associations between any gene and immunity (e.g., Lymphocytes, Immunomodulators and Chemokines) are pre-calculated for 30 TCGA cancers.
2.8. Linked Omics Database
To explore the biological function of CLEC4A and its role in LUAD, an analysis was performed using the LinkedOmics database. LinkedOmic allows the analysis of a wide range of cancers. The LinkFinder module screens for CLEC4A-associated differentially expressed genes in LUAD and presents them through volcanoes and heatmaps. Multi-omics and pan-cancer analysis, Gene Ontology (GO) analysis and Genome Encyclopedia (KEGG) pathway enrichment analysis are performed in the LinInterpreter module. (http://www.linkedomics.org). GO analysis includes biological processes (BP), cellular components (CC) and molecular functions (MF).
2.9. Analysis of the Relationship between CLEC4A Expression and Clinicopathological Characteristics
Logistic regression and Wilcoxon signed rank test were used to analyse the relationship between CLEC4A expression and clinicopathological characteristics: age, gender, smoking, clinical stage, T stage, M stage, N stage and primary therapy outcome.
2.10. Prognostic Analysis of CLEC4A
To investigate whether prognostic characteristics could be independent of other clinical parameters (including age, gender, smoking, and pathological stage), univariate and multivariate Cox regression models were performed on TCGA-LUAD data. P<0.05 was considered to be statistically significant, and variables with P<0.05 in multivariate Cox analysis were identified as independent prognostic factors for LUAD. In addition, to verify the correlation between CLEC4A expression and survival in LUAD, the effect of CLEC4A on overall survival (OS), progression-free survival (PFS), and disease-specific survival (DSS) in LUAD patients was assessed using the Kaplan-Meier method. The relationship between CLEC4A expression and OS in clinical subgroups of LUAD was also analysed. The analysis could not be complete for each clinical parameter as clinical information was not complete for some samples, and there were differences in the total number of samples for some different clinical parameters.
2.11. Statistical Analysis
All data in this paper are presented as mean ± standard deviation (SD). The R programming language is used for this study, and all survival analyses are performed using the R package 'survival', with Pearson correlation coefficients used for all correlation analyses. P < 0.05 is considered statistically significant and is expressed as follows:* P < 0.05, ** P < 0.01 and *** P < 0.001.
3. RESULTS
3.1. mRNA Expression Levels of CLEC4A in Different Cancers
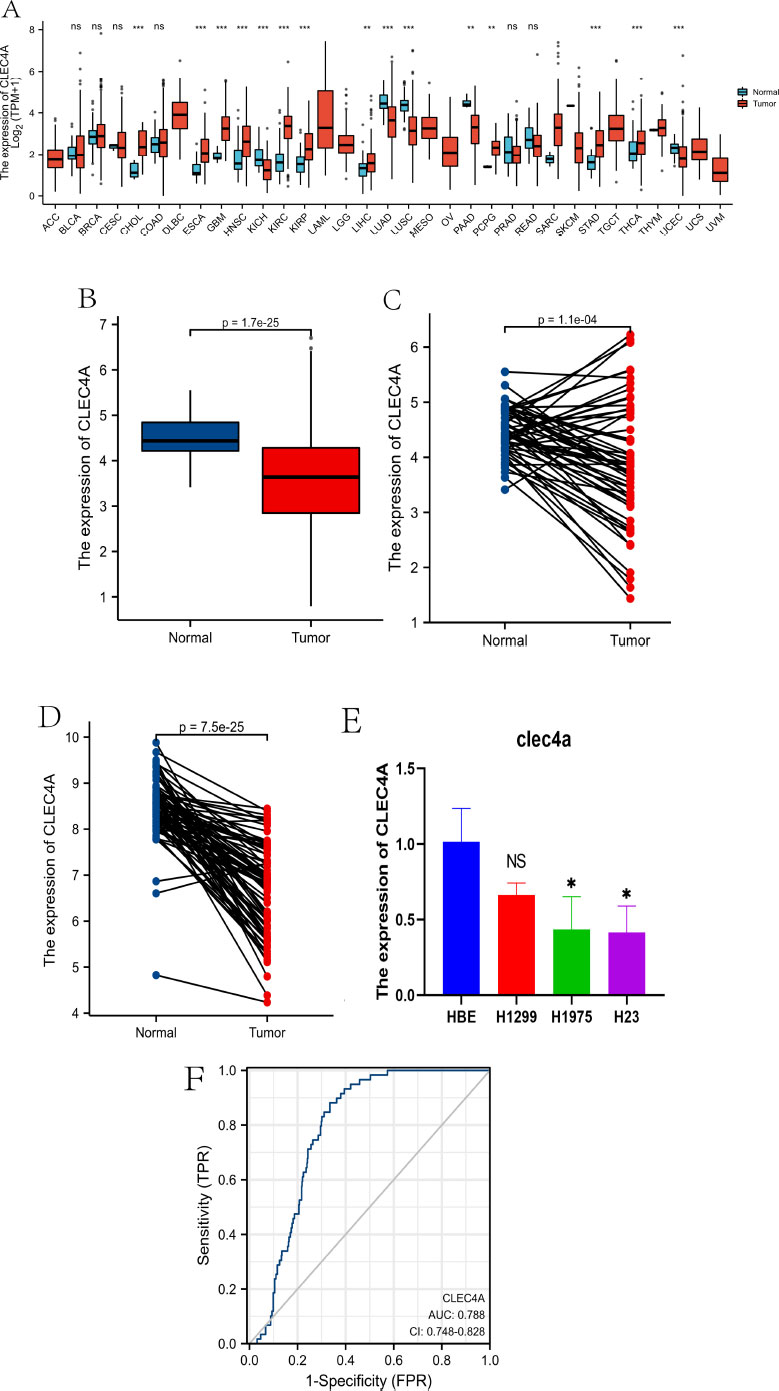
In order to gain further insights into the relationship between CLEC4A and cancer, we conducted a comprehensive analysis of 33 different tumor types using the TCGA dataset. Our findings, as depicted in Fig. (1A), highlight the distinct expression patterns of CLEC4A in numerous tumor types and their corresponding para- cancerous tissues. We observed significant upregulation of CLEC4A in several cancers, such as Bladder Urothelial Carcinoma (BLCA), Breast invasive carcinoma (BRCA), Cholangio- carcinoma (CHOL), Colon adenocarcinoma (COAD), Esophageal carcinoma (ESCA), Glioblastoma multiforme (GBM), Head and Neck squamous cell carcinoma (HNSC), Kidney renal clear cell carcinoma (KIRC), Kidney renal papillary cell carcinoma (KIRP), Liver hepatocellular carcinoma (LIHC), Pheochromocytoma and Paraganglioma (PCPG), Stomach adenocarcinoma (STAD), and Thyroid carcinoma (THCA). Conversely, in Cervical squamous cell carcinoma and endocervical adeno- carcinoma (CESC), Kidney Chromophobe (KICH), Lung adenocarcinoma (LUAD), Lung squamous cell carcinoma (LUSC), Pancreatic adenocarcinoma (PAAD), Prostate adenocarcinoma (PRAD), Rectum adenocarcinoma (READ), and Uterine Corpus Endometrial Carcinoma (UCEC), the expression of CLEC4A was found to be downregulated in tumor tissues compared to adjacent normal tissues. To further investigate the role of CLEC4A specifically in LUAD, we analyzed the expression levels of CLEC4A in a substantial sample set comprising 535 LUAD samples and 59 paraneoplastic samples from the TCGA database. Our analysis consistently revealed lower expression of CLEC4A in normal lung tissues compared to LUAD tissues, as shown in Fig. (1A-C). These findings were validated using the independent GSE75037 dataset, which includes 83 matched pairs of lung adenocarcinomas and non-malignant adjacent tissue, as shown in Fig. (1D). To assess the diagnostic potential of CLEC4A in distinguishing LUAD from normal lung tissue, we generated ROC curves using the TCGA internal test set. Fig. (1F) illustrates that CLEC4A exhibits a high discriminatory power, with an area under the curve (AUC) value of 0.788. This indicates that CLEC4A may serve as a reliable diagnostic biomarker for LUAD.
In order to gain mechanistic insights into the role of CLEC4A in LUAD development, we performed qRT-PCR analysis on various lung adenocarcinoma cell lines, including H1975 and H23, as well as the normal lung cell line 16HBE. The results, presented in Fig. (1E), demonstrate significantly lower relative mRNA levels of CLEC4A in H1975 (p-value <0.05) and H23 (p-value <0.05) cell lines, while no significant downregulation was observed in the H1299 cell line (p-value > 0.05) compared to 16HBE. These findings provide additional evidence supporting the potential regulatory role of CLEC4A in LUAD development. In summary, our comprehensive analysis of CLEC4A expression in various tumors, including LUAD, highlights its differential expression patterns and establishes its potential diagnostic value in distinguishing tumor tissues from normal tissues. The downregulation of CLEC4A in certain tumor types, including LUAD, suggests its involvement in tumor development. These findings position CLEC4A as a promising target for further exploration in the field of LUAD research, offering potential diagnostic and therapeutic value.
3.2. Relationship between CLEC4A Expression and Clinicopathological Features in LUAD Patients
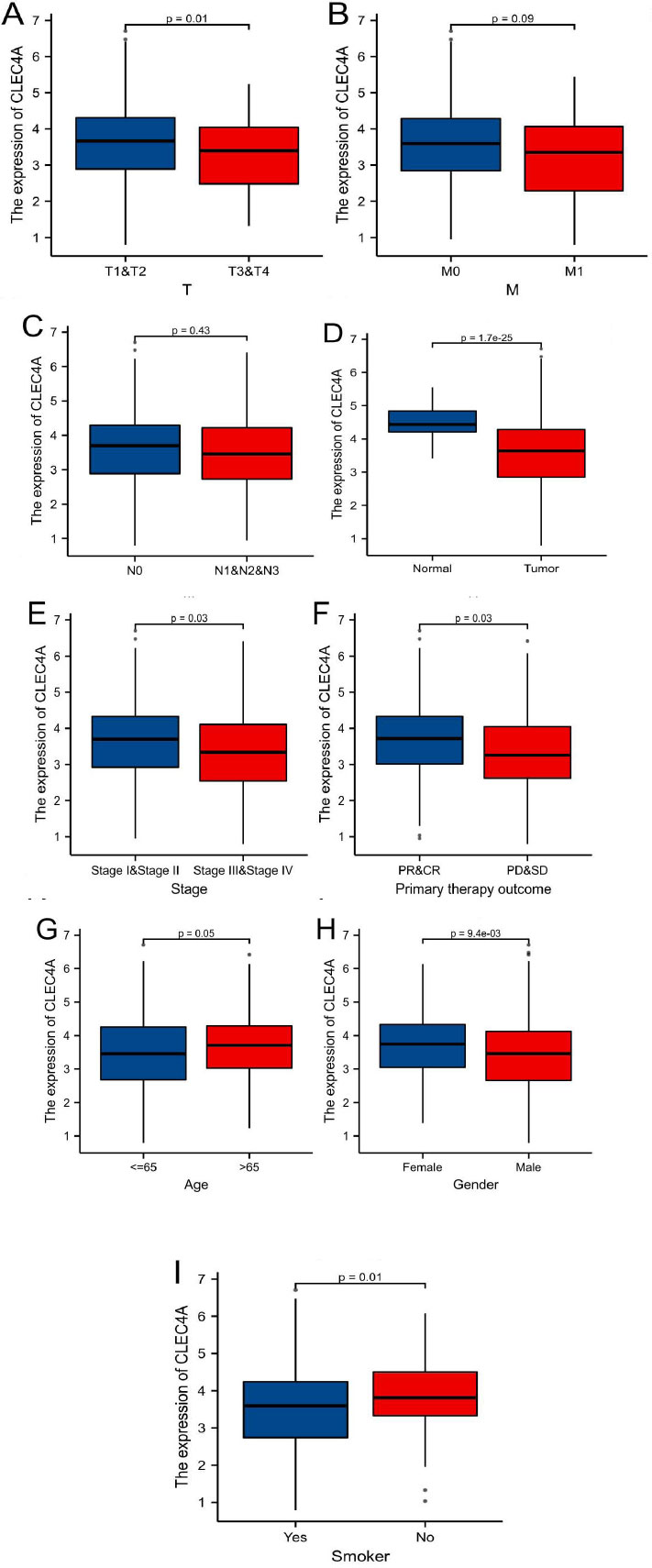
To conduct our study, we collected data from RNA-seq analysis and clinical records of 533 patients diagnosed with lung adenocarcinoma (LUAD) from the TCGA database (refer to Table 1 for details). The LUAD cohort was divided into two groups, namely the high expression group and the low expression group, based on the median values of CLEC4A expression. We examined the correlation between CLEC4A expression and various clinical features using the Wilcoxon signed rank test (Fig. 2A-I). Through our investigation of the correlation between CLEC4A expression and various pathological cancer stages, we found that the downregulation of CLEC4A is associated with the advanced stages of lung adenocarcinoma. Our analysis revealed significant associations between CLEC4A expression and different clinical parameters. Specifically, low CLEC4A expression was found to be positively correlated with a higher grade of T stage (p=0.01). We can speculate that CLEC4A may achieve its anti-tumor effects by limiting the rate of in situ tumor growth. Furthermore, pathological CLEC4A expression demonstrated significant associations with tumor status (P<0.001), gender (P<0.01), smoking history (P=0.01), age (P=0.05), and response to immunotherapy (P=0.03). These findings highlight the potential role of CLEC4A expression as a prognostic indicator and suggest its relevance in predicting various clinicopathological characteristics of LUAD patients. Moreover, our study utilized univariate logistic regression analysis to further explore the relationship between CLEC4A expression and specific clinical features (Table 2). The results reveal a robust association between CLEC4A and gender, with an odds ratio (OR) of 0.603 (95% confidence interval: 0.428-0.849) and a p-value of 0.004. Additionally, we observed a significant correlation between CLEC4A expression and primary treatment effect, with an OR of 1.967 (95% confidence interval: 1.273-3.097) and a p-value of 0.003. These findings suggest that CLEC4A expression may serve as a predictive biomarker for gender-related differences in LUAD as well as treatment response. These findings contribute to a better understanding of the role of CLEC4A and its potential implications in LUAD management, allowing for the development of targeted therapeutic strategies and personalized treatment approaches.
| Characteristic | levels | Low expression of CLEC4A | High expression of CLEC4A | p |
|---|---|---|---|---|
| n | - | 267 | 268 | - |
| T stage, n (%) | T1 | 78 (14.7%) | 97 (18.2%) | 0.225 |
| T2 | 148 (27.8%) | 141 (26.5%) | - | |
| T3 | 29 (5.5%) | 20 (3.8%) | - | |
| T4 | 11 (2.1%) | 8 (1.5%) | - | |
| N stage, n (%) | N0 | 166 (32%) | 182 (35.1%) | 0.352 |
| N1 | 52 (10%) | 43 (8.3%) | - | |
| N2 | 42 (8.1%) | 32 (6.2%) | - | |
| N3 | 1 (0.2%) | 1 (0.2%) | - | |
| M stage, n (%) | M0 | 186 (48.2%) | 175 (45.3%) | 0.821 |
| M1 | 14 (3.6%) | 11 (2.8%) | - | |
| Gender, n (%) | Female | 126 (23.6%) | 160 (29.9%) | 0.005 |
| Male | 141 (26.4%) | 108 (20.2%) | - | |
| Age, n (%) | <=65 | 138 (26.7%) | 117 (22.7%) | 0.078 |
| >65 | 120 (23.3%) | 141 (27.3%) | - | |
| Residual tumor, n (%) | R0 | 182 (48.9%) | 173 (46.5%) | 0.101 |
| R1 | 8 (2.2%) | 5 (1.3%) | - | |
| R2 | 0 (0%) | 4 (1.1%) | - | |
| Primary therapy outcome, n (%) | PD | 43 (9.6%) | 28 (6.3%) | 0.02 |
| SD | 24 (5.4%) | 13 (2.9%) | - | |
| PR | 3 (0.7%) | 3 (0.7%) | - | |
| CR | 150 (33.6%) | 182 (40.8%) | - | |
| Smoker, n (%) | No | 32 (6.1%) | 43 (8.3%) | 0.193 |
| Yes | 230 (44.1%) | 216 (41.5%) | - | |
| OS event, n (%) | Alive | 149 (27.9%) | 194 (36.3%) | < 0.001 |
| Dead | 118 (22.1%) | 74 (13.8%) | - | |
| Age, median (IQR) | - | 64.5 (58, 72) | 67 (59, 73) | 0.079 |
3.3. Prognostic Value of CLEC4A Expression in LUAD
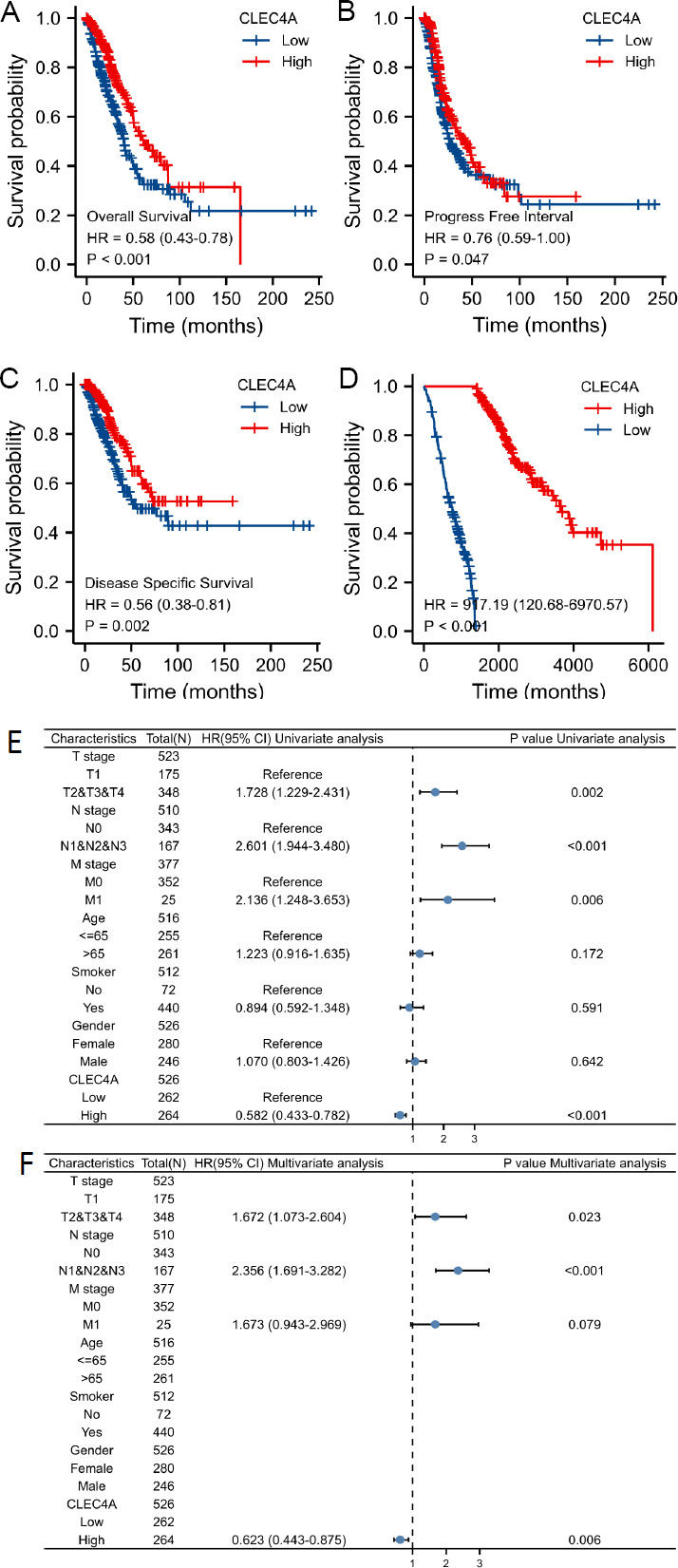
To verify the prognostic significance of CLEC4A expression in lung adenocarcinoma (LUAD), we conducted an analysis using data from the TCGA and GEO datasets through the Kaplan-Meier Plotter database. High expression levels of CLEC4A were associated with poor prognosis in LUAD patients, as illustrated in Fig. (3A). To account for potential biases arising from non-cancer-related events, we further examined progression-free survival (PFS) and disease-specific survival (DSS) (refer to Fig. 3B-3C). Remarkably, the results for PFS and DSS mirrored those for overall survival (OS), indicating that lower CLEC4A expression was correlated with improved PFS and DSS outcomes. To validate our findings, we employed the GSE68465 dataset as an independent cohort (refer to Fig. 3D). The results were consistent with the TCGA findings, affirming that CLEC4A expression significantly impacts the prognosis of LUAD patients. Furthermore, we utilized Cox proportional hazard regression models to analyze various prognostic factors. Through univariate and multivariate analysis of clinical characteristics (as shown in Fig. 3E-3F), we discovered that CLEC4A expression (HR=1.605, P=0.006), advanced T-stage (HR=1.672, P=0.023), and positive N-stage (HR=2.356, P<0.001) served as independent prognostic factors. These results highlight the potential of CLEC4A expression as a valuable indicator specific to the T-stage and N-stage status, offering significant insights into the prognosis of LUAD patients. Collectively, our comprehensive analysis using the Kaplan-Meier Plotter database, along with validation in an independent dataset, establishes the prognostic value of CLEC4A expression in LUAD. High expression levels of CLEC4A are associated with poor prognosis, while lower expression correlates with improved outcomes in terms of PFS, DSS, and overall survival. Moreover, our Cox regression analysis identifies CLEC4A expression, along with advanced T-stage and positive N-stage, as independent prognostic factors. These findings underscore the significance of CLEC4A as a potential marker for assessing LUAD prognosis and provide valuable insights for personalized treatment strategies and patient management.
| Characteristics | Total(N) | Odds Ratio(OR) | P value |
|---|---|---|---|
| T stage (T2&T3&T4 vs. T1) | 532 | 0.723 (0.502-1.039) | 0.08 |
| N stage (N1&N2&N3 vs. N0) | 519 | 0.730 (0.504-1.053) | 0.093 |
| M stage (M1 vs. M0) | 386 | 0.835 (0.361-1.884) | 0.665 |
| Gender (Male vs. Female) | 535 | 0.603 (0.428-0.849) | 0.004 |
| Pathologic stage (Stage III&Stage IV vs. Stage I&Stage II) | 527 | 0.688 (0.448-1.048) | 0.083 |
| Primary therapy outcome (PR&CR vs. PD&SD) | 446 | 1.976 (1.273-3.097) | 0.003 |
| Age (>65 vs. <=65) | 516 | 1.386 (0.981-1.961) | 0.065 |
| Smoker (Yes vs. No) | 521 | 0.699 (0.424-1.142) | 0.155 |
3.4. Predictive Value of CLEC4A Levels based on Clinical Subgroups
In order to further substantiate the predictive significance of CLEC4A in lung adenocarcinoma (LUAD), we sought to explore the relationship between CLEC4A expression and overall survival (OS) among diverse subgroups defined by specific clinical characteristics. Our analysis unveiled compelling findings, indicating that LUAD patients displaying higher levels of CLEC4A expression exhibited improved OS outcomes across several subgroups. Notably, within the Stage I-II subgroup (Fig. 4A), high CLEC4A expression was associated with better OS outcomes. Similarly, in subgroups characterized by T1-T2 stage tumors (Fig. 4B), absence of distant metastasis (M0) (Fig. 4C), male gender (Fig. 4D), age greater than 65 years (Fig. 4E), and White race (Fig. 4F), higher CLEC4A expression levels were consistently correlated with enhanced OS in LUAD patients. These findings signify the potential utility of CLEC4A as a prognostic biomarker in various clinical contexts. The association between elevated CLEC4A expression and improved OS across different subgroups highlights the extensive clinically relevant predictive value of this molecular marker in LUAD. By distinguishing patients with a favorable prognosis, CLEC4A expression assess- ment may aid in treatment planning and therapeutic decision-making, enabling personalized approaches for individuals who may benefit most from specific inter- ventions. Our analysis demonstrates the positive cor- relation between high CLEC4A expression and better OS outcomes not only in the overall LUAD population but also within subgroups stratified by crucial clinical character- istics such as disease stage, tumor size, metastasis status, gender, age, and race. These findings underscore the potential of CLEC4A as a robust predictive indicator, paving the way for its integration into clinical practice to aid in outcome prediction and optimize patient manage- ment strategies in LUAD.
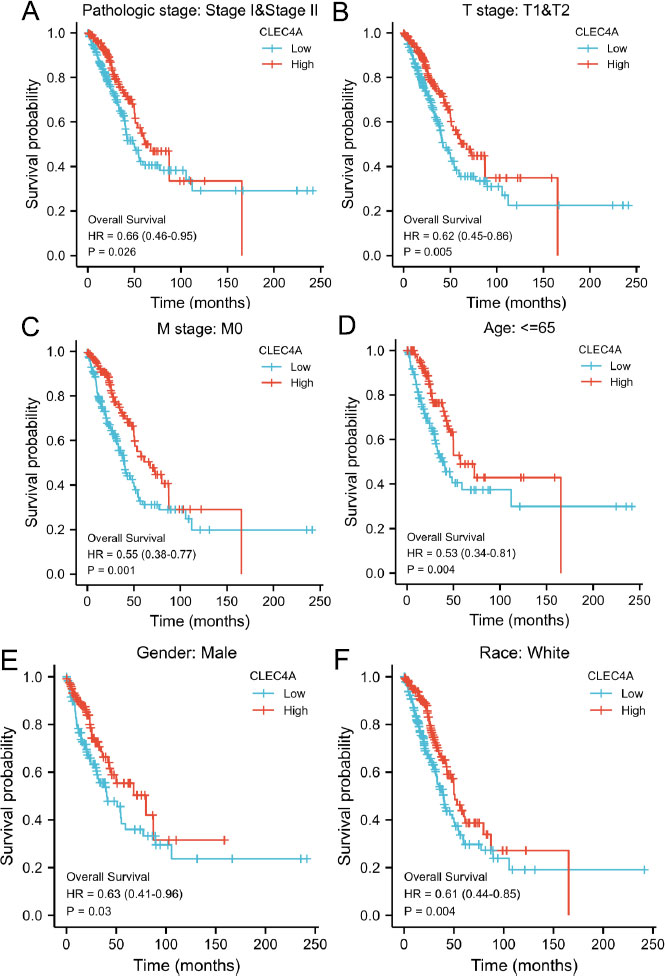 |
Fig. (4). Association of CLEC4A expression levels in TCGA with OS in different clinical subgroups of LUAD, including stage I-II (A), T1-T2 (B), M0 (C), and age<65. |
3.5. Co-expression Network of CLEC4A Expression in LUAD
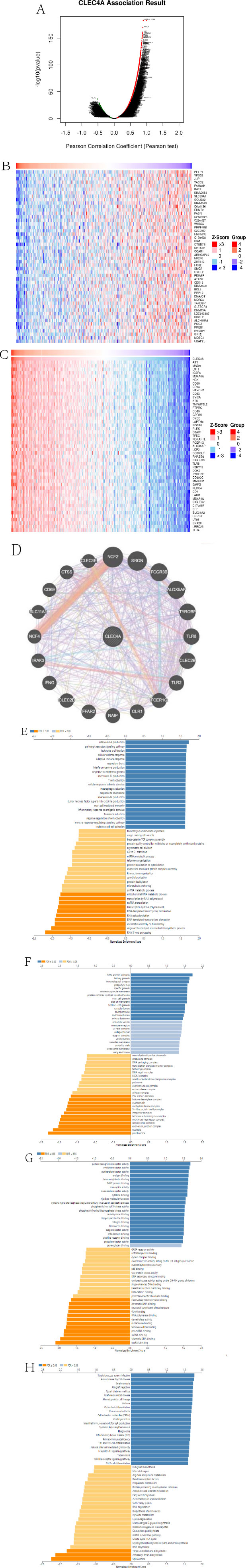
In order to gain insight into the role of CLEC4A in LUAD, a comprehensive analysis was conducted using the LinkedOmics database. A total of 9,430 genes (depicted as dark green dots) and 10,558 positively associated genes (depicted as dark red dots) that exhibited a negative correlation with CLEC4A expression were examined (Fig. 5A and Table S1). To further investigate the association between CLEC4A expression and specific genes, heatmaps for the top 50 positively and negatively associated genes with CLEC4A expression were generated and presented in Fig. (5B and 5C), respectively. One of the main objectives of this study was to identify the significant genes associated with CLEC4A. This was accomplished by constructing a PPI network, which revealed the top 20 genes that exhibited a significant connection with CLEC4A (Fig. 5D). To gain deeper insights into the biological function of CLEC4A and its associated genes, the LinkInterpreter module in the LinkedOmics portal was utilized for GO-BP, GO-CC, GO-MF, and KEGG analysis (Fig. 5E-H).
The GO term annotation analysis provides valuable information regarding the biological processes, cellular components, and molecular functions associated with CLEC4A co-expressed genes. The results indicated that these genes primarily contribute to interleukin-4 production, purinergic receptor signaling pathway, leukocyte proliferation, T cell activation, tumor necrosis factor superfamily cytokine production, MHC protein complex, tertiary granule formation, immunological synapse formation, pattern recognition receptor activity, and cytokine receptor activity. Furthermore, the KEGG pathway analysis revealed the significance of CLEC4A co-expressed genes in various pathological conditions. These genes were found to play crucial roles in Staphylococcus aureus infection, autoimmune thyroid disease, Leishmaniasis, Type I rejection, allograft rejection, Type I diabetes mellitus, graft-versus-host disease, and hematopoietic cell lineage regulation. Based on the comprehensive analysis conducted, it can be inferred that the CLEC4A expression network modulates autoimmune responses and hampers tumor proliferation during cancer development, thereby influencing the prognosis of LUAD. These findings shed light on the potential importance of CLEC4A as a therapeutic target in the context of LUAD.
3.6. Relationship between CLEC4A and Immune Molecules
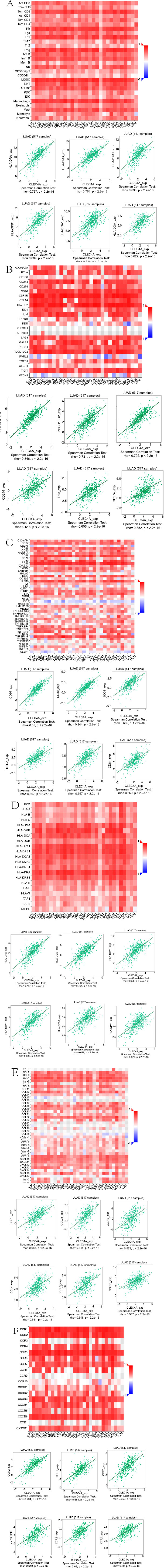
To enhance our comprehension of CLEC4A and its impact on the immune microenvironment, an extensive investigation was conducted using the TISIDB database. This analysis aimed to explore the correlation between CLEC4A and several key immunological features, such as tumor-infiltrating lymphocytes (TIL), immunomodulators, chemokines, and receptors. Immunomodulators were categorized into three groups: immunosuppressive agents, immunostimulants, and major histocompatibility complex (MHC) molecules. The results shown in Fig. (6A), unveiled the six TILs with the highest relevance to CLEC4A, specifically Treg_abundance, MDSC_abundance, Macrophage_abundance, Tfh_abundance, Act_DC_abun dance, and Th1_abundance. Fig. (6B) presents the six immunoinhibitors that exhibited the strongest correlation with CLEC4A, namely HAVCR2_exp, CSF1R_exp, PDCD1LG2_exp, CD244_exp, IL-10_exp, and CD274_exp.
Conversely, Fig. (6C) demonstrates the six immuno- stimulators with the highest CLEC4A correlation, including CD86_exp, CD80_exp, ICOS_exp, IL2RA_exp, CD48_exp, and CD28_exp, indicating that elevated CLEC4A expression was associated with a more robust immune response. Furthermore, our analysis revealed the six MHC molecules (HLA-DRA_exp, HLA-DMB_exp, HLA-DPA1_exp, HLA-DPB1_exp, HLA-DQA1_exp, and HLA-DOA_exp) that exhibited the strongest correlation with CLEC4A, as shown in Fig. (6D). These findings suggest that increased CLEC4A expression is linked to a heightened immune response. Additionally, Fig. (6E) showcases the six chemokines (CCL13_exp, CCL23_exp, CCL17_exp, CCL4_exp, CCL3_exp, and CCL18_exp) that displayed the highest correlation with CLEC4A. Similarly, Fig. (6F) presents the six receptors (CCR2_exp, CCR1_exp, CCR5_exp, CCR6_exp, CCR8_exp, and CCR4_exp) exhibiting the strongest correlation with CLEC4A. Taken together, our study provides compelling evidence that CLEC4A plays an active role in the regulation of various immune molecules in LUAD. Moreover, it influences immune infiltration within the tumor microenvironment. These findings emphasize the significance of CLEC4A in modulating immune responses and may pave the way for targeted therapeutic strategies in the context of LUAD.
3.7. Relationship between CLEC4A Expression and Immune Infiltration
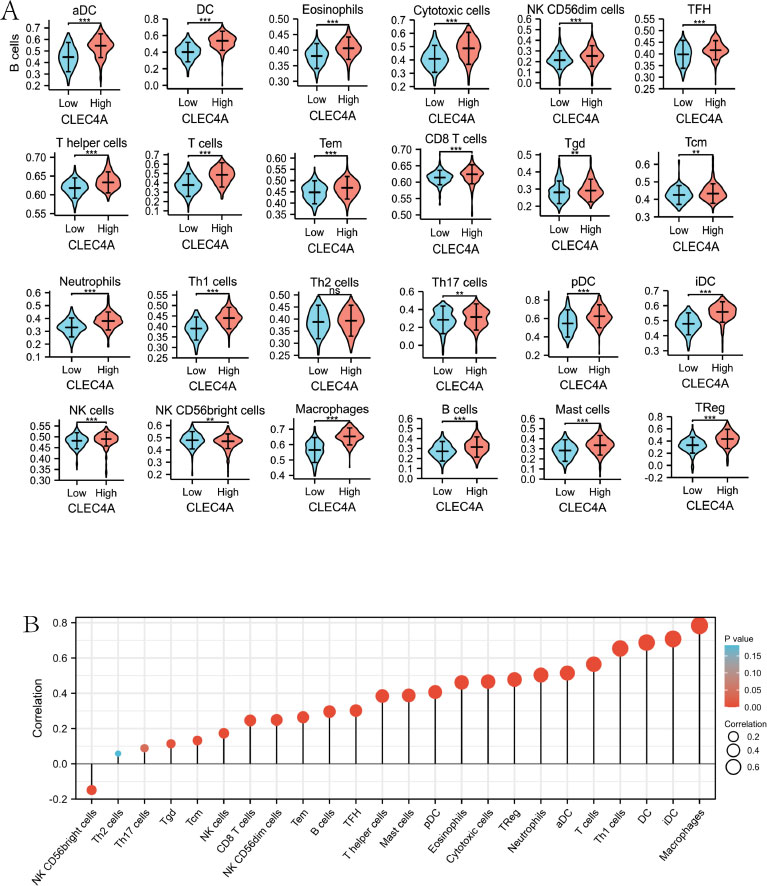
In order to gain insights into the connection between CLEC4A expression and immune cell infiltrates, an in-depth analysis was performed to evaluate the correlation between CLEC4A expression and 24 distinct immune cell types in LUAD. The findings of this analysis revealed a robust positive correlation between CLEC4A expression levels and nearly all immune cell infiltrates, including T cells, T helper cells, B cells, NK cells, CD8+ T cells, and various others. Intriguingly, a negative correlation was observed with NK CD56bright cells, as depicted in Fig. (7A). Moreover, the analysis demonstrated significant disparities in immune cell infiltration between the high and low CLEC4A expression groups. Remarkably, the high-expression group exhibited a substantially higher rate of immune cell infiltration compared to the low-expression group, as illustrated in Fig. (7B). These
findings underscore the direct association between CLEC4A expression and the presence of immune cell infiltrates in LUAD. The strong positive correlation observed between CLEC4A expression and diverse immune cell types suggests a potential role for CLEC4A in regulating immune responses within the tumor microenvironment. Furthermore, the disparities in immune cell infiltration rates between the high and low CLEC4A expression groups indicate that CLEC4A expression levels may impact the immune landscape of LUAD tumors. Understanding the relationship between CLEC4A and immune cell infiltration is crucial in uncovering the underlying mechanisms that contribute to tumor progression and may provide valuable insights for the development of immunotherapeutic strategies targeting CLEC4A in the treatment of LUAD. Further research is warranted to elucidate the precise role of CLEC4A in immune cell modulation and its potential implications for therapeutic interventions.
3.8. Mutations between High and Low CLEC4A Groups in LUAD
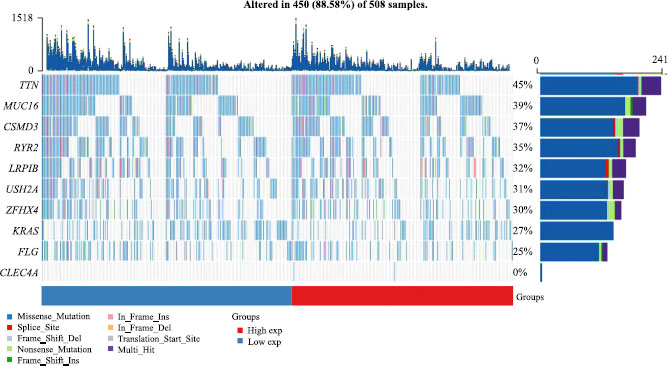
Understanding the role of gene mutations in promoting tumor development and metastasis is crucial for advancing our knowledge of oncology and developing targeted therapies. To investigate the impact of gene mutations on LUAD, particularly in relation to CLEC4A expression, we conducted a comprehensive analysis at the genomic level. Specifically, LUAD patients were divided into two groups based on the median CLEC4A expression values, and a comparison was made regarding the differences in mutations between these two groups. The analysis yielded intriguing results, demonstrating that the frequency of mutations was significantly higher in patients with high CLEC4A expression compared to those with low CLEC4A expression, as indicated in Fig. (8). This finding suggests that high CLEC4A expression may contribute to the improvement of the tumor microenvironment and facilitate disease progression in patients with LUAD.
4. DISCUSSION
Lung adenocarcinoma (LUAD) places a significant financial burden on families despite gradual improvements in treatments that enhance human survival. Unfortunately, a poor prognosis continues to affect patients. LUAD tumors exhibit a high level of heterogeneity, and genes can undergo thousands of mutations, including copy number variation (CNA), simple nucleotide variation (SNV), and somatic structural variation (SSV). Furthermore, dysregulation of the microenvironment, oncogenic drivers, and genetic mutations can significantly impact tumor behavior [11]. One notable characteristic of LUAD is the release of a complex array of inflammatory signals by tumor cells. This leads to the recruitment and infiltration of cytokines, chemokines, growth factors, tumor cells, and various immune cells, resulting in a diverse and intricate tumor microenvironment (TME). In many cases, the TME serves as the primary cause of immunosuppression, enabling tumor progression through several mechanisms. These mechanisms include the inhibition of T cell activation and cytotoxicity, promotion of the conversion of macrophages into suppressive tumor-associated macrophages (TAM), recruitment of regulatory T cells (Tregs) and infiltration of myeloid-derived suppressor cells (MDSC), as well as the secretion of suppressive cytokines and metabolites. In response to the challenges posed by tumor immunosuppression, immune checkpoint inhibitor therapy has been developed and clinically implemented in the treatment of non-small cell lung cancer (NSCLC). Currently, monoclonal antibodies (mAb) targeting ligands of programmed cell death protein (PD-1) or the cytotoxic T lymphocyte-associated protein 4 (CTLA-4) pathway serve as approved immune checkpoint blockers [12, 13]. Therefore, the future of cancer immunotherapy appears promising. Nonetheless, the effectiveness of cancer immunotherapy varies significantly among individuals due to the lack of accurate prediction of the predominant patient population. Consequently, there is an urgent need to develop novel LUAD biomarkers to enable the administration of precise therapies to patients [14].
The CLR (C-type lectin receptor) has gained significant attention in recent years for its role in regulating both innate and adaptive immunity. Cells of the innate immune response, such as macrophages, dendritic cells (DCs), neutrophils, and phagocytose extracellular pathogens or virus-infected host cells. They then present pathogen-associated proteins to T cells, activating the adaptive immune system [15, 16]. DCs, being the most potent antigen-presenting cells, play a critical role in regulating the functions of both B and T lymphocytes. They are also involved in controlling T-cell tolerance to reduce autoimmune responses [17]. The CLR serves as a pattern recognition receptor (PRR) and recognizes carbohydrate ligands found on infected microbes. Binding to these ligands triggers multiple signaling cascades, including NF-κB, through immunoreceptor tyrosine-based activation motifs (ITAM) or ITAM in the Fc receptor gamma chain (FcRγ) [18]. NF-κB is involved in regulating Treg (regulatory T cell) development and function, cell proliferation, apoptosis, and gene expression for lymphocyte activation [19]. One specific CLR that mediates T-cell responses is DCIR (CLEC4A) [20]. In humans, there is a single gene encoding human DCIR (hDCIR), whereas in mice, there are four DCIR-like proteins (mDCIR1, mDCIR2, mDCIR3, mDCIR4). Human CLEC4A is differentially expressed by DCs. Mature DCs capture antigens and present them to naive T cells, initiating an immune response. On the other hand, immature DCs promote Treg function, leading to immunosuppression [21-23]. CLEC4A belongs to the Dectin-2 family of C-type lectin-like receptors [24]. Unlike other members of the Dectin-2 family, only hDCIR, mDCIR1, and mDCIR2 contain ITIM (immunoreceptor tyrosine-based inhibitory motif) [25, 26]. hDCIR and the ITIM of mDCIR1 activate the phosphatases SHP-1 and SHP-2, mediating inhibitory signaling [27-29]. In vitro studies targeting antigens to DCIR have demonstrated cross-presentation of DCs, Langerhans cells, blood marrow type DCs, and plasma cell type DCs to CD8+ T cells, promoting immune responses in humans. A study by Weng et al. used CLEC4A shRNA in a mouse bladder tumor model showed that regulation of CLEC4A induced anti-tumor responses and cellular immunity in mice [30]. This indicates the potential therapeutic value of targeting CLEC4A in cancer immunotherapy.
Recent studies have illustrated the tremendous potential of C-type lectin receptors (CLRs) in the realm of immunotherapy. However, limited investigations have focused on understanding the significance of CLEC4A in lung adenocarcinoma (LUAD) and its potential as a diagnostic and therapeutic target. Hence, the objective of this study was to fill this research gap by utilizing high-throughput RNA sequencing data retrieved from the TCGA database, followed by bioinformatics analysis. Our findings based on these analyses revealed a reduction in CLEC4A expression in eight different types of cancer, namely Cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), Kidney Chromophobe (KICH), LUAD, Lung squamous cell carcinoma (LUSC), Pancreatic adenocarcinoma (PAAD), Prostate adenocarcinoma (PRAD), Rectum adenocarcinoma (READ), and Uterine Corpus Endometrial Carcinoma (UCEC). In the context of LUAD, the decreased expression of CLEC4A corresponded to the progression of the tumor, including the expansion of the primary tumor extent, the appearance of distant metastases, and poor treatment outcomes. Remarkably, the diagnosis of LUAD displayed an area under the receiver operating characteristic curve (AUC) value of 0.788, suggesting that CLEC4A potentially serves as a valuable diagnostic marker for this disease. Furthermore, reduced CLEC4A expression indicated primary treatment outcome, age, advanced T-stage, survival time, and a dismal prognosis. These findings emphasize that CLEC4A has prognostic value as a biomarker for LUAD, with low expression levels being associated with disease progression and a poor prognosis.
Another crucial aspect of our study entailed investigating the genes associated with CLEC4A in LUAD, as well as the pathways and functions enriched by CLEC4A. Consequently, Gene Ontology (GO) enrichment analysis highlighted several predominant biological functions linked to CLEC4A, including interleukin-4 production, leukocyte proliferation, T cell activation, tumor necrosis factor superfamily cytokine production, and major histocompatibility complex protein complex. To delve into the immunological mechanisms at play, we examined the correlation between CLEC4A expression and the extent of immune cell infiltration in LUAD. Encouragingly, we observed that high CLEC4A expression correlated with increased infiltration of numerous immune cell types, including B cells, natural killer (NK) cells, CD8+ T cells, and notably, T cells and dendritic cells (DC). Additionally, CLEC4A exhibited active involvement in the regulation of various immune molecules.
However, we must acknowledge some limitations of the current study. Firstly, most of the data analysis was based on public databases, and the samples used in our study were predominantly collected retrospectively. Therefore, inherent case selection bias may have an impact on the results. To validate our findings, extensive prospective studies, as well as in vivo and in vitro experiments, are needed. Although our study was analyzed and validated using many patient samples from different databases, it opened new perspectives and outlooks for cancer treatment. Based on this study, researchers could aim to understand the outstanding potential of CLEC4A in tumor immunity, conduct various experiments to explore it in-depth and contribute to cancer treatment.
CONCLUSION
In conclusion, our research strongly suggests that CLEC4A acts as a critical regulator in suppressing LUAD onset and progression. The significant correlation between high CLEC4A expression and various immune-related characteristics in LUAD emphasizes its potential as a valuable biomarker and therapeutic target associated with LUAD patient prognosis. Nonetheless, further experi- mental validations are necessary to establish the precise relationship between CLEC4A expression and LUAD development. This study not only enhances our understanding of the anti-tumor mechanisms involving CLEC4A but also presents new strategies for improving the clinical management of LUAD patients.
LIST OF ABBREVIATIONS
| CLEC | = C-type (Ca2+-dependent) Lectin |
| LUAD | = Lung Adenocarcinoma |
| DC | = Dendritic Cells |
ETHICAL STATEMENT
The Ethics Committee of the Affiliated Hospital of Nantong University has granted exemptions from approval for research related to the use of such public databases.
CONSENT FOR PUBLICATION
Not applicable.
AVAILABILITY OF DATA AND MATERIALS
The data of this manuscript can be downloaded from The Cancer Genome Atlas database (https://portal.gdc. cancer.gov/) and the GEO database (https://www.ncbi. nlm.nih.gov/geo/).
FUNDING
This work has been funded with support from the Nantong Science and Technology Plan Project (Grant Nos. JC22022023) and Nantong Municipal Health Commission (Grant Nos. QN2023008).
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
ACKNOWLEDGEMENTS
Declared none.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s website along with the published article. The data supporting the findings of the article are available in the Zenodo repository at URL, reference number 10.5281/zenodo.10656502.